Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Research paper
- How to Write a Discussion Section | Tips & Examples

How to Write a Discussion Section | Tips & Examples
Published on August 21, 2022 by Shona McCombes . Revised on July 18, 2023.

The discussion section is where you delve into the meaning, importance, and relevance of your results .
It should focus on explaining and evaluating what you found, showing how it relates to your literature review and paper or dissertation topic , and making an argument in support of your overall conclusion. It should not be a second results section.
There are different ways to write this section, but you can focus your writing around these key elements:
- Summary : A brief recap of your key results
- Interpretations: What do your results mean?
- Implications: Why do your results matter?
- Limitations: What can’t your results tell us?
- Recommendations: Avenues for further studies or analyses
Instantly correct all language mistakes in your text
Upload your document to correct all your mistakes in minutes
Table of contents
What not to include in your discussion section, step 1: summarize your key findings, step 2: give your interpretations, step 3: discuss the implications, step 4: acknowledge the limitations, step 5: share your recommendations, discussion section example, other interesting articles, frequently asked questions about discussion sections.
There are a few common mistakes to avoid when writing the discussion section of your paper.
- Don’t introduce new results: You should only discuss the data that you have already reported in your results section .
- Don’t make inflated claims: Avoid overinterpretation and speculation that isn’t directly supported by your data.
- Don’t undermine your research: The discussion of limitations should aim to strengthen your credibility, not emphasize weaknesses or failures.
Don't submit your assignments before you do this
The academic proofreading tool has been trained on 1000s of academic texts. Making it the most accurate and reliable proofreading tool for students. Free citation check included.
Try for free
Start this section by reiterating your research problem and concisely summarizing your major findings. To speed up the process you can use a summarizer to quickly get an overview of all important findings. Don’t just repeat all the data you have already reported—aim for a clear statement of the overall result that directly answers your main research question . This should be no more than one paragraph.
Many students struggle with the differences between a discussion section and a results section . The crux of the matter is that your results sections should present your results, and your discussion section should subjectively evaluate them. Try not to blend elements of these two sections, in order to keep your paper sharp.
- The results indicate that…
- The study demonstrates a correlation between…
- This analysis supports the theory that…
- The data suggest that…
The meaning of your results may seem obvious to you, but it’s important to spell out their significance for your reader, showing exactly how they answer your research question.
The form of your interpretations will depend on the type of research, but some typical approaches to interpreting the data include:
- Identifying correlations , patterns, and relationships among the data
- Discussing whether the results met your expectations or supported your hypotheses
- Contextualizing your findings within previous research and theory
- Explaining unexpected results and evaluating their significance
- Considering possible alternative explanations and making an argument for your position
You can organize your discussion around key themes, hypotheses, or research questions, following the same structure as your results section. Alternatively, you can also begin by highlighting the most significant or unexpected results.
- In line with the hypothesis…
- Contrary to the hypothesized association…
- The results contradict the claims of Smith (2022) that…
- The results might suggest that x . However, based on the findings of similar studies, a more plausible explanation is y .
As well as giving your own interpretations, make sure to relate your results back to the scholarly work that you surveyed in the literature review . The discussion should show how your findings fit with existing knowledge, what new insights they contribute, and what consequences they have for theory or practice.
Ask yourself these questions:
- Do your results support or challenge existing theories? If they support existing theories, what new information do they contribute? If they challenge existing theories, why do you think that is?
- Are there any practical implications?
Your overall aim is to show the reader exactly what your research has contributed, and why they should care.
- These results build on existing evidence of…
- The results do not fit with the theory that…
- The experiment provides a new insight into the relationship between…
- These results should be taken into account when considering how to…
- The data contribute a clearer understanding of…
- While previous research has focused on x , these results demonstrate that y .
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
Even the best research has its limitations. Acknowledging these is important to demonstrate your credibility. Limitations aren’t about listing your errors, but about providing an accurate picture of what can and cannot be concluded from your study.
Limitations might be due to your overall research design, specific methodological choices , or unanticipated obstacles that emerged during your research process.
Here are a few common possibilities:
- If your sample size was small or limited to a specific group of people, explain how generalizability is limited.
- If you encountered problems when gathering or analyzing data, explain how these influenced the results.
- If there are potential confounding variables that you were unable to control, acknowledge the effect these may have had.
After noting the limitations, you can reiterate why the results are nonetheless valid for the purpose of answering your research question.
- The generalizability of the results is limited by…
- The reliability of these data is impacted by…
- Due to the lack of data on x , the results cannot confirm…
- The methodological choices were constrained by…
- It is beyond the scope of this study to…
Based on the discussion of your results, you can make recommendations for practical implementation or further research. Sometimes, the recommendations are saved for the conclusion .
Suggestions for further research can lead directly from the limitations. Don’t just state that more studies should be done—give concrete ideas for how future work can build on areas that your own research was unable to address.
- Further research is needed to establish…
- Future studies should take into account…
- Avenues for future research include…

If you want to know more about AI for academic writing, AI tools, or research bias, make sure to check out some of our other articles with explanations and examples or go directly to our tools!
Research bias
- Anchoring bias
- Halo effect
- The Baader–Meinhof phenomenon
- The placebo effect
- Nonresponse bias
- Deep learning
- Generative AI
- Machine learning
- Reinforcement learning
- Supervised vs. unsupervised learning
(AI) Tools
- Grammar Checker
- Paraphrasing Tool
- Text Summarizer
- AI Detector
- Plagiarism Checker
- Citation Generator
In the discussion , you explore the meaning and relevance of your research results , explaining how they fit with existing research and theory. Discuss:
- Your interpretations : what do the results tell us?
- The implications : why do the results matter?
- The limitation s : what can’t the results tell us?
The results chapter or section simply and objectively reports what you found, without speculating on why you found these results. The discussion interprets the meaning of the results, puts them in context, and explains why they matter.
In qualitative research , results and discussion are sometimes combined. But in quantitative research , it’s considered important to separate the objective results from your interpretation of them.
In a thesis or dissertation, the discussion is an in-depth exploration of the results, going into detail about the meaning of your findings and citing relevant sources to put them in context.
The conclusion is more shorter and more general: it concisely answers your main research question and makes recommendations based on your overall findings.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
McCombes, S. (2023, July 18). How to Write a Discussion Section | Tips & Examples. Scribbr. Retrieved June 26, 2024, from https://www.scribbr.com/dissertation/discussion/
Is this article helpful?
Shona McCombes
Other students also liked, how to write a literature review | guide, examples, & templates, what is a research methodology | steps & tips, how to write a results section | tips & examples, get unlimited documents corrected.
✔ Free APA citation check included ✔ Unlimited document corrections ✔ Specialized in correcting academic texts
- Research Process
- Manuscript Preparation
- Manuscript Review
- Publication Process
- Publication Recognition
- Language Editing Services
- Translation Services

6 Steps to Write an Excellent Discussion in Your Manuscript
- 4 minute read
Table of Contents
The discussion section in scientific manuscripts might be the last few paragraphs, but its role goes far beyond wrapping up. It’s the part of an article where scientists talk about what they found and what it means, where raw data turns into meaningful insights. Therefore, discussion is a vital component of the article.
An excellent discussion is well-organized. We bring to you authors a classic 6-step method for writing discussion sections, with examples to illustrate the functions and specific writing logic of each step. Take a look at how you can impress journal reviewers with a concise and focused discussion section!
Discussion frame structure
Conventionally, a discussion section has three parts: an introductory paragraph, a few intermediate paragraphs, and a conclusion¹. Please follow the steps below:
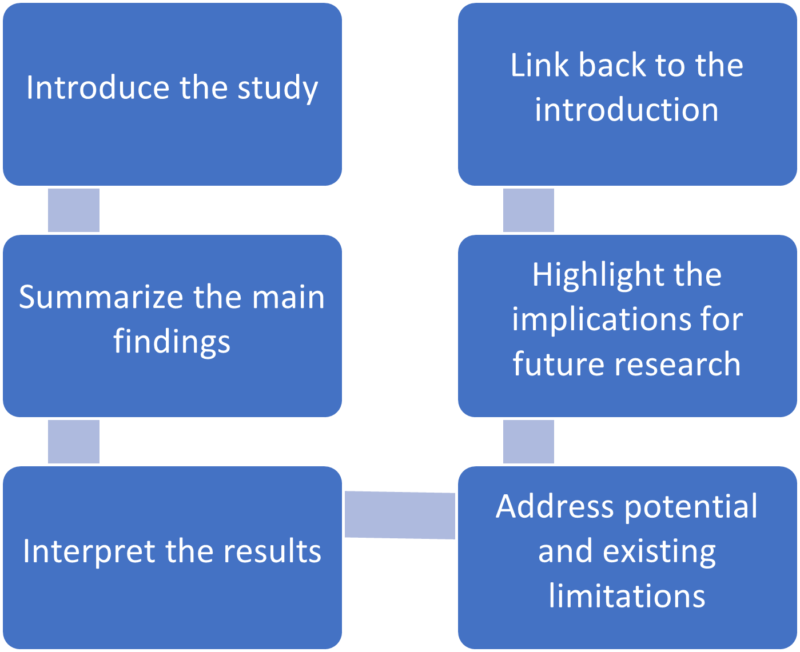
1.Introduction—mention gaps in previous research¹⁻ ²
Here, you orient the reader to your study. In the first paragraph, it is advisable to mention the research gap your paper addresses.
Example: This study investigated the cognitive effects of a meat-only diet on adults. While earlier studies have explored the impact of a carnivorous diet on physical attributes and agility, they have not explicitly addressed its influence on cognitively intense tasks involving memory and reasoning.
2. Summarizing key findings—let your data speak ¹⁻ ²
After you have laid out the context for your study, recapitulate some of its key findings. Also, highlight key data and evidence supporting these findings.
Example: We found that risk-taking behavior among teenagers correlates with their tendency to invest in cryptocurrencies. Risk takers in this study, as measured by the Cambridge Gambling Task, tended to have an inordinately higher proportion of their savings invested as crypto coins.
3. Interpreting results—compare with other papers¹⁻²
Here, you must analyze and interpret any results concerning the research question or hypothesis. How do the key findings of your study help verify or disprove the hypothesis? What practical relevance does your discovery have?
Example: Our study suggests that higher daily caffeine intake is not associated with poor performance in major sporting events. Athletes may benefit from the cardiovascular benefits of daily caffeine intake without adversely impacting performance.
Remember, unlike the results section, the discussion ideally focuses on locating your findings in the larger body of existing research. Hence, compare your results with those of other peer-reviewed papers.
Example: Although Miller et al. (2020) found evidence of such political bias in a multicultural population, our findings suggest that the bias is weak or virtually non-existent among politically active citizens.
4. Addressing limitations—their potential impact on the results¹⁻²
Discuss the potential impact of limitations on the results. Most studies have limitations, and it is crucial to acknowledge them in the intermediary paragraphs of the discussion section. Limitations may include low sample size, suspected interference or noise in data, low effect size, etc.
Example: This study explored a comprehensive list of adverse effects associated with the novel drug ‘X’. However, long-term studies may be needed to confirm its safety, especially regarding major cardiac events.
5. Implications for future research—how to explore further¹⁻²
Locate areas of your research where more investigation is needed. Concluding paragraphs of the discussion can explain what research will likely confirm your results or identify knowledge gaps your study left unaddressed.
Example: Our study demonstrates that roads paved with the plastic-infused compound ‘Y’ are more resilient than asphalt. Future studies may explore economically feasible ways of producing compound Y in bulk.
6. Conclusion—summarize content¹⁻²
A good way to wind up the discussion section is by revisiting the research question mentioned in your introduction. Sign off by expressing the main findings of your study.
Example: Recent observations suggest that the fish ‘Z’ is moving upriver in many parts of the Amazon basin. Our findings provide conclusive evidence that this phenomenon is associated with rising sea levels and climate change, not due to elevated numbers of invasive predators.
A rigorous and concise discussion section is one of the keys to achieving an excellent paper. It serves as a critical platform for researchers to interpret and connect their findings with the broader scientific context. By detailing the results, carefully comparing them with existing research, and explaining the limitations of this study, you can effectively help reviewers and readers understand the entire research article more comprehensively and deeply¹⁻² , thereby helping your manuscript to be successfully published and gain wider dissemination.
In addition to keeping this writing guide, you can also use Elsevier Language Services to improve the quality of your paper more deeply and comprehensively. We have a professional editing team covering multiple disciplines. With our profound disciplinary background and rich polishing experience, we can significantly optimize all paper modules including the discussion, effectively improve the fluency and rigor of your articles, and make your scientific research results consistent, with its value reflected more clearly. We are always committed to ensuring the quality of papers according to the standards of top journals, improving the publishing efficiency of scientific researchers, and helping you on the road to academic success. Check us out here !
Type in wordcount for Standard Total: USD EUR JPY Follow this link if your manuscript is longer than 12,000 words. Upload
References:
- Masic, I. (2018). How to write an efficient discussion? Medical Archives , 72(3), 306. https://doi.org/10.5455/medarh.2018.72.306-307
- Şanlı, Ö., Erdem, S., & Tefik, T. (2014). How to write a discussion section? Urology Research & Practice , 39(1), 20–24. https://doi.org/10.5152/tud.2013.049

Navigating “Chinglish” Errors in Academic English Writing

A Guide to Crafting Shorter, Impactful Sentences in Academic Writing
You may also like.

Page-Turner Articles are More Than Just Good Arguments: Be Mindful of Tone and Structure!

A Must-see for Researchers! How to Ensure Inclusivity in Your Scientific Writing

Make Hook, Line, and Sinker: The Art of Crafting Engaging Introductions

Can Describing Study Limitations Improve the Quality of Your Paper?

How to Write Clear and Crisp Civil Engineering Papers? Here are 5 Key Tips to Consider

The Clear Path to An Impactful Paper: ②

The Essentials of Writing to Communicate Research in Medicine
Input your search keywords and press Enter.
How to Write the Discussion Section of a Research Paper
The discussion section of a research paper analyzes and interprets the findings, provides context, compares them with previous studies, identifies limitations, and suggests future research directions.
Updated on September 15, 2023

Structure your discussion section right, and you’ll be cited more often while doing a greater service to the scientific community. So, what actually goes into the discussion section? And how do you write it?
The discussion section of your research paper is where you let the reader know how your study is positioned in the literature, what to take away from your paper, and how your work helps them. It can also include your conclusions and suggestions for future studies.
First, we’ll define all the parts of your discussion paper, and then look into how to write a strong, effective discussion section for your paper or manuscript.
Discussion section: what is it, what it does
The discussion section comes later in your paper, following the introduction, methods, and results. The discussion sets up your study’s conclusions. Its main goals are to present, interpret, and provide a context for your results.
What is it?
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research.
This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study (introduction), how you did it (methods), and what happened (results). In the discussion, you’ll help the reader connect the ideas from these sections.
Why is it necessary?
The discussion provides context and interpretations for the results. It also answers the questions posed in the introduction. While the results section describes your findings, the discussion explains what they say. This is also where you can describe the impact or implications of your research.
Adds context for your results
Most research studies aim to answer a question, replicate a finding, or address limitations in the literature. These goals are first described in the introduction. However, in the discussion section, the author can refer back to them to explain how the study's objective was achieved.
Shows what your results actually mean and real-world implications
The discussion can also describe the effect of your findings on research or practice. How are your results significant for readers, other researchers, or policymakers?
What to include in your discussion (in the correct order)
A complete and effective discussion section should at least touch on the points described below.
Summary of key findings
The discussion should begin with a brief factual summary of the results. Concisely overview the main results you obtained.
Begin with key findings with supporting evidence
Your results section described a list of findings, but what message do they send when you look at them all together?
Your findings were detailed in the results section, so there’s no need to repeat them here, but do provide at least a few highlights. This will help refresh the reader’s memory and help them focus on the big picture.
Read the first paragraph of the discussion section in this article (PDF) for an example of how to start this part of your paper. Notice how the authors break down their results and follow each description sentence with an explanation of why each finding is relevant.
State clearly and concisely
Following a clear and direct writing style is especially important in the discussion section. After all, this is where you will make some of the most impactful points in your paper. While the results section often contains technical vocabulary, such as statistical terms, the discussion section lets you describe your findings more clearly.
Interpretation of results
Once you’ve given your reader an overview of your results, you need to interpret those results. In other words, what do your results mean? Discuss the findings’ implications and significance in relation to your research question or hypothesis.
Analyze and interpret your findings
Look into your findings and explore what’s behind them or what may have caused them. If your introduction cited theories or studies that could explain your findings, use these sources as a basis to discuss your results.
For example, look at the second paragraph in the discussion section of this article on waggling honey bees. Here, the authors explore their results based on information from the literature.
Unexpected or contradictory results
Sometimes, your findings are not what you expect. Here’s where you describe this and try to find a reason for it. Could it be because of the method you used? Does it have something to do with the variables analyzed? Comparing your methods with those of other similar studies can help with this task.
Context and comparison with previous work
Refer to related studies to place your research in a larger context and the literature. Compare and contrast your findings with existing literature, highlighting similarities, differences, and/or contradictions.
How your work compares or contrasts with previous work
Studies with similar findings to yours can be cited to show the strength of your findings. Information from these studies can also be used to help explain your results. Differences between your findings and others in the literature can also be discussed here.
How to divide this section into subsections
If you have more than one objective in your study or many key findings, you can dedicate a separate section to each of these. Here’s an example of this approach. You can see that the discussion section is divided into topics and even has a separate heading for each of them.
Limitations
Many journals require you to include the limitations of your study in the discussion. Even if they don’t, there are good reasons to mention these in your paper.
Why limitations don’t have a negative connotation
A study’s limitations are points to be improved upon in future research. While some of these may be flaws in your method, many may be due to factors you couldn’t predict.
Examples include time constraints or small sample sizes. Pointing this out will help future researchers avoid or address these issues. This part of the discussion can also include any attempts you have made to reduce the impact of these limitations, as in this study .
How limitations add to a researcher's credibility
Pointing out the limitations of your study demonstrates transparency. It also shows that you know your methods well and can conduct a critical assessment of them.
Implications and significance
The final paragraph of the discussion section should contain the take-home messages for your study. It can also cite the “strong points” of your study, to contrast with the limitations section.
Restate your hypothesis
Remind the reader what your hypothesis was before you conducted the study.
How was it proven or disproven?
Identify your main findings and describe how they relate to your hypothesis.
How your results contribute to the literature
Were you able to answer your research question? Or address a gap in the literature?
Future implications of your research
Describe the impact that your results may have on the topic of study. Your results may show, for instance, that there are still limitations in the literature for future studies to address. There may be a need for studies that extend your findings in a specific way. You also may need additional research to corroborate your findings.
Sample discussion section
This fictitious example covers all the aspects discussed above. Your actual discussion section will probably be much longer, but you can read this to get an idea of everything your discussion should cover.
Our results showed that the presence of cats in a household is associated with higher levels of perceived happiness by its human occupants. These findings support our hypothesis and demonstrate the association between pet ownership and well-being.
The present findings align with those of Bao and Schreer (2016) and Hardie et al. (2023), who observed greater life satisfaction in pet owners relative to non-owners. Although the present study did not directly evaluate life satisfaction, this factor may explain the association between happiness and cat ownership observed in our sample.
Our findings must be interpreted in light of some limitations, such as the focus on cat ownership only rather than pets as a whole. This may limit the generalizability of our results.
Nevertheless, this study had several strengths. These include its strict exclusion criteria and use of a standardized assessment instrument to investigate the relationships between pets and owners. These attributes bolster the accuracy of our results and reduce the influence of confounding factors, increasing the strength of our conclusions. Future studies may examine the factors that mediate the association between pet ownership and happiness to better comprehend this phenomenon.
This brief discussion begins with a quick summary of the results and hypothesis. The next paragraph cites previous research and compares its findings to those of this study. Information from previous studies is also used to help interpret the findings. After discussing the results of the study, some limitations are pointed out. The paper also explains why these limitations may influence the interpretation of results. Then, final conclusions are drawn based on the study, and directions for future research are suggested.
How to make your discussion flow naturally
If you find writing in scientific English challenging, the discussion and conclusions are often the hardest parts of the paper to write. That’s because you’re not just listing up studies, methods, and outcomes. You’re actually expressing your thoughts and interpretations in words.
- How formal should it be?
- What words should you use, or not use?
- How do you meet strict word limits, or make it longer and more informative?
Always give it your best, but sometimes a helping hand can, well, help. Getting a professional edit can help clarify your work’s importance while improving the English used to explain it. When readers know the value of your work, they’ll cite it. We’ll assign your study to an expert editor knowledgeable in your area of research. Their work will clarify your discussion, helping it to tell your story. Find out more about AJE Editing.

Adam Goulston, PsyD, MS, MBA, MISD, ELS
Science Marketing Consultant
See our "Privacy Policy"
Ensure your structure and ideas are consistent and clearly communicated
Pair your Premium Editing with our add-on service Presubmission Review for an overall assessment of your manuscript.
Training videos | Faqs

Discussion Section Examples and Writing Tips
Abstract | Introduction | Literature Review | Research question | Materials & Methods | Results | Discussion | Conclusion
In this blog, we look at how to write the discussion section of a research paper. We will go through plenty of discussion examples and understand how to construct a great discussion section for your research paper.
1. What is the purpose of the discussion section?
The discussion section is one of the most important sections of your research paper. This is where you interpret your results, highlight your contributions, and explain the value of your work to your readers. This is one of the challenging parts to write because the author must clearly explain the significance of their results and tie everything back to the research questions.
2. How should I structure my discussion section?
Generally, the discussion section of a research paper typically contains the following parts.
Research summary It is a good idea to start this section with an overall summary of your work and highlight the main findings of your research.
Interpretation of findings You must interpret your findings clearly to your readers one by one.
Comparison with literature You must talk about how your results fit into existing research in the literature.
Implications of your work You should talk about the implications and possible benefits of your research.
Limitations You should talk about the possible limitations and shortcomings of your research
Future work And finally, you can talk about the possible future directions of your work.
3. Discussion Examples
Let’s look at some examples of the discussion section. We will be looking at discussion examples from different fields and of different formats. We have split this section into multiple components so that it is easy for you to digest and understand.
3.1. An example of research summary in discussion
It is a good idea to start your discussion section with the summary of your work. The best way to do this will be to restate your research question, and then reminding your readers about your methods, and finally providing an overall summary of your results.
Our aims were to compare the effectiveness and user-friendliness of different storm detection software for storm tracking. On the basis of these aims, we ran multiple experiments with the same conditions using different storm detection software. Our results showed that in both speed and accuracy of data, ‘software A’ performed better than ‘software B’. _ Aims summary _ Methodology summary _ Results summary
This discussion example is from an engineering research paper. The authors are restating their aims first, which is to compare different types of storm-tracking software. Then, they are providing a brief summary of the methods. Here, they are testing different storm-tracking software under different conditions to see which performs the best. Then, they are finally providing their main finding which is that they found ‘software A’ better than ‘software B’. This is a very good example of how to start the discussion section by presenting a summary of your work.
3.2. An example of result interpretation in discussion
The next step is to interpret your results. You have to explain your results clearly to your readers. Here is a discussion example that shows how to interpret your results.
The results of this study indicate significant differences between classical music and pop music in terms of their effects on memory recall and cognition. This implies that as the complexity of the music increases, so does its ability to facilitate cognitive processing. This finding aligns with the well-known “Mozart effect,” which suggests that listening to classical music can enhance cognitive function. _ Result _ Interpretation _ Additional evidence
The authors are saying that their results show that there is a significant difference between pop music and classical music in terms of memory recall and cognition. Now they are providing their interpretation of the findings. They think it is because there is a link between the complexity of music and cognitive processing. They are also making a reference to a well-known theory called the ‘Mozart effect’ to back up their findings. It is a nicely written passage and the author’s interpretation sounds very convincing and credible.
3.3. An example of literature comparison in discussion
The next step is to compare your results to the literature. You have to explain clearly how your findings compare with similar findings made by other researchers. Here is a discussion example where authors are providing details of papers in the literature that both support and oppose their findings.
Our analysis predicts that climate change will have a significant impact on wheat yield. This finding undermines one of the central pieces of evidence in some previous simulation studies [1-3] that suggest a negative effect of climate change on wheat yield, but the result is entirely consistent with the predictions of other research [4-5] that suggests the overall change in climate could result in increases in wheat yield. _ Result _ Comparison with literature
The authors are saying that their results show that climate change will have a significant effect on wheat production. Then, they are saying that there are some papers in the literature that are in agreement with their findings. However, there are also many papers in the literature that disagree with their findings. This is very important. Your discussion should be two-sided, not one-sided. You should not ignore the literature that doesn’t corroborate your findings.
3.4. An example of research implications in discussion
The next step is to explain to your readers how your findings will benefit society and the research community. You have to clearly explain the value of your work to your readers. Here is a discussion example where authors explain the implications of their research.
The results contribute insights with regard to the management of wildfire events using artificial intelligence. One could easily argue that the obvious practical implication of this study is that it proposes utilizing cloud-based machine vision to detect wildfires in real-time, even before the first responders receive emergency calls. _ Your finding _ Implications of your finding
In this paper, the authors are saying that their findings indicate that Artificial intelligence can be used to effectively manage wildfire events. Then, they are talking about the practical implications of their study. They are saying that their work has proven that machine learning can be used to detect wildfires in real-time. This is a great practical application and can save thousands of lives. As you can see, after reading this passage, you can immediately understand the value and significance of the work.
3.5. An example of limitations in discussion
It is very important that you discuss the limitations of your study. Limitations are flaws and shortcomings of your study. You have to tell your readers how your limitations might influence the outcomes and conclusions of your research. Most studies will have some form of limitation. So be honest and don’t hide your limitations. In reality, your readers and reviewers will be impressed with your paper if you are upfront about your limitations.
Study design and small sample size are important limitations. This could have led to an overestimation of the effect. Future research should reconfirm these findings by conducting larger-scale studies. _ Limitation _ How it might affect the results? _ How to fix the limitation?
Here is a discussion example where the author talks about study limitations. The authors are saying that the main limitations of the study are the small sample size and weak study design. Then they explain how this might have affected their results. They are saying that it is possible that they are overestimating the actual effect they are measuring. Then finally they are telling the readers that more studies with larger sample sizes should be conducted to reconfirm the findings.
As you can see, the authors are clearly explaining three things here:
3.6. An example of future work in discussion
It is important to remember not to end your paper with limitations. Finish your paper on a positive note by telling your readers about the benefits of your research and possible future directions. Here is a discussion example where the author talks about future work.
Our study highlights useful insights about the potential of biomass as a renewable energy source. Future research can extend this research in several ways, including research on how to tackle challenges that hinder the sustainability of renewable energy sources towards climate change mitigation, such as market failures, lack of information and access to raw materials. _ Benefits of your work _ Future work
The authors are starting the final paragraph of the discussion section by highlighting the benefit of their work which is the use of biomass as a renewable source of energy. Then they talk about future research. They are saying that future research can focus on how to improve the sustainability of biomass production. This is a very good example of how to finish the discussion section of your paper on a positive note.
4. Frequently Asked Questions
Sometimes you will have negative or unexpected results in your paper. You have to talk about it in your discussion section. A lot of students find it difficult to write this part. The best way to handle this situation is not to look at results as either positive or negative. A result is a result, and you will always have something important and interesting to say about your findings. Just spend some time investigating what might have caused this result and tell your readers about it.
You must talk about the limitations of your work in the discussion section of the paper. One of the important qualities that the scientific community expects from a researcher is honesty and admitting when they have made a mistake. The important trick you have to learn while presenting your limitations is to present them in a constructive way rather than being too negative about them. You must try to use positive language even when you are talking about major limitations of your work.
If you have something exciting to say about your results or found something new that nobody else has found before, then, don’t be modest and use flat language when presenting this in the discussion. Use words like ‘break through’, ‘indisputable evidence’, ‘exciting proposition’ to increase the impact of your findings.
Important thing to remember is not to overstate your findings. If you found something really interesting but are not 100% sure, you must not mislead your readers. The best way to do this will be to use words like ‘it appears’ and ‘it seems’. This will tell the readers that there is a slight possibility that you might be wrong.
Similar Posts

Figures and Tables in Research Papers – Tips and Examples
In this blog, we will look at best practices for presenting tables and figures in your research paper.

Critical Literature Review : How to Critique a Research Article?
In this blog, we will look at how to use constructive language when critiquing other’s work in your research paper.

Introduction Paragraph Examples and Writing Tips
In this blog, we will go through a few introduction paragraph examples and understand how to construct a great introduction paragraph for your research paper.

Materials and Methods Examples and Writing Tips
In this blog, we will go through many materials and methods examples and understand how to write a clear and concise method section for your research paper.

3 Costly Mistakes to Avoid in the Research Introduction
In this blog, we will discuss three common mistakes that beginner writers make while writing the research paper introduction.

Technical Terms, Notations, and Scientific Jargon in Research Papers
In this blog, we will teach you how to use specialized terminology in your research papers with some practical examples.
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- 5 Share Facebook
- 7 Share Twitter
- 5 Share LinkedIn
- 10 Share Email
- USC Libraries
- Research Guides
Organizing Your Social Sciences Research Paper
- 8. The Discussion
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The purpose of the discussion section is to interpret and describe the significance of your findings in relation to what was already known about the research problem being investigated and to explain any new understanding or insights that emerged as a result of your research. The discussion will always connect to the introduction by way of the research questions or hypotheses you posed and the literature you reviewed, but the discussion does not simply repeat or rearrange the first parts of your paper; the discussion clearly explains how your study advanced the reader's understanding of the research problem from where you left them at the end of your review of prior research.
Annesley, Thomas M. “The Discussion Section: Your Closing Argument.” Clinical Chemistry 56 (November 2010): 1671-1674; Peacock, Matthew. “Communicative Moves in the Discussion Section of Research Articles.” System 30 (December 2002): 479-497.
Importance of a Good Discussion
The discussion section is often considered the most important part of your research paper because it:
- Most effectively demonstrates your ability as a researcher to think critically about an issue, to develop creative solutions to problems based upon a logical synthesis of the findings, and to formulate a deeper, more profound understanding of the research problem under investigation;
- Presents the underlying meaning of your research, notes possible implications in other areas of study, and explores possible improvements that can be made in order to further develop the concerns of your research;
- Highlights the importance of your study and how it can contribute to understanding the research problem within the field of study;
- Presents how the findings from your study revealed and helped fill gaps in the literature that had not been previously exposed or adequately described; and,
- Engages the reader in thinking critically about issues based on an evidence-based interpretation of findings; it is not governed strictly by objective reporting of information.
Annesley Thomas M. “The Discussion Section: Your Closing Argument.” Clinical Chemistry 56 (November 2010): 1671-1674; Bitchener, John and Helen Basturkmen. “Perceptions of the Difficulties of Postgraduate L2 Thesis Students Writing the Discussion Section.” Journal of English for Academic Purposes 5 (January 2006): 4-18; Kretchmer, Paul. Fourteen Steps to Writing an Effective Discussion Section. San Francisco Edit, 2003-2008.
Structure and Writing Style
I. General Rules
These are the general rules you should adopt when composing your discussion of the results :
- Do not be verbose or repetitive; be concise and make your points clearly
- Avoid the use of jargon or undefined technical language
- Follow a logical stream of thought; in general, interpret and discuss the significance of your findings in the same sequence you described them in your results section [a notable exception is to begin by highlighting an unexpected result or a finding that can grab the reader's attention]
- Use the present verb tense, especially for established facts; however, refer to specific works or prior studies in the past tense
- If needed, use subheadings to help organize your discussion or to categorize your interpretations into themes
II. The Content
The content of the discussion section of your paper most often includes :
- Explanation of results : Comment on whether or not the results were expected for each set of findings; go into greater depth to explain findings that were unexpected or especially profound. If appropriate, note any unusual or unanticipated patterns or trends that emerged from your results and explain their meaning in relation to the research problem.
- References to previous research : Either compare your results with the findings from other studies or use the studies to support a claim. This can include re-visiting key sources already cited in your literature review section, or, save them to cite later in the discussion section if they are more important to compare with your results instead of being a part of the general literature review of prior research used to provide context and background information. Note that you can make this decision to highlight specific studies after you have begun writing the discussion section.
- Deduction : A claim for how the results can be applied more generally. For example, describing lessons learned, proposing recommendations that can help improve a situation, or highlighting best practices.
- Hypothesis : A more general claim or possible conclusion arising from the results [which may be proved or disproved in subsequent research]. This can be framed as new research questions that emerged as a consequence of your analysis.
III. Organization and Structure
Keep the following sequential points in mind as you organize and write the discussion section of your paper:
- Think of your discussion as an inverted pyramid. Organize the discussion from the general to the specific, linking your findings to the literature, then to theory, then to practice [if appropriate].
- Use the same key terms, narrative style, and verb tense [present] that you used when describing the research problem in your introduction.
- Begin by briefly re-stating the research problem you were investigating and answer all of the research questions underpinning the problem that you posed in the introduction.
- Describe the patterns, principles, and relationships shown by each major findings and place them in proper perspective. The sequence of this information is important; first state the answer, then the relevant results, then cite the work of others. If appropriate, refer the reader to a figure or table to help enhance the interpretation of the data [either within the text or as an appendix].
- Regardless of where it's mentioned, a good discussion section includes analysis of any unexpected findings. This part of the discussion should begin with a description of the unanticipated finding, followed by a brief interpretation as to why you believe it appeared and, if necessary, its possible significance in relation to the overall study. If more than one unexpected finding emerged during the study, describe each of them in the order they appeared as you gathered or analyzed the data. As noted, the exception to discussing findings in the same order you described them in the results section would be to begin by highlighting the implications of a particularly unexpected or significant finding that emerged from the study, followed by a discussion of the remaining findings.
- Before concluding the discussion, identify potential limitations and weaknesses if you do not plan to do so in the conclusion of the paper. Comment on their relative importance in relation to your overall interpretation of the results and, if necessary, note how they may affect the validity of your findings. Avoid using an apologetic tone; however, be honest and self-critical [e.g., in retrospect, had you included a particular question in a survey instrument, additional data could have been revealed].
- The discussion section should end with a concise summary of the principal implications of the findings regardless of their significance. Give a brief explanation about why you believe the findings and conclusions of your study are important and how they support broader knowledge or understanding of the research problem. This can be followed by any recommendations for further research. However, do not offer recommendations which could have been easily addressed within the study. This would demonstrate to the reader that you have inadequately examined and interpreted the data.
IV. Overall Objectives
The objectives of your discussion section should include the following: I. Reiterate the Research Problem/State the Major Findings
Briefly reiterate the research problem or problems you are investigating and the methods you used to investigate them, then move quickly to describe the major findings of the study. You should write a direct, declarative, and succinct proclamation of the study results, usually in one paragraph.
II. Explain the Meaning of the Findings and Why They are Important
No one has thought as long and hard about your study as you have. Systematically explain the underlying meaning of your findings and state why you believe they are significant. After reading the discussion section, you want the reader to think critically about the results and why they are important. You don’t want to force the reader to go through the paper multiple times to figure out what it all means. If applicable, begin this part of the section by repeating what you consider to be your most significant or unanticipated finding first, then systematically review each finding. Otherwise, follow the general order you reported the findings presented in the results section.
III. Relate the Findings to Similar Studies
No study in the social sciences is so novel or possesses such a restricted focus that it has absolutely no relation to previously published research. The discussion section should relate your results to those found in other studies, particularly if questions raised from prior studies served as the motivation for your research. This is important because comparing and contrasting the findings of other studies helps to support the overall importance of your results and it highlights how and in what ways your study differs from other research about the topic. Note that any significant or unanticipated finding is often because there was no prior research to indicate the finding could occur. If there is prior research to indicate this, you need to explain why it was significant or unanticipated. IV. Consider Alternative Explanations of the Findings
It is important to remember that the purpose of research in the social sciences is to discover and not to prove . When writing the discussion section, you should carefully consider all possible explanations for the study results, rather than just those that fit your hypothesis or prior assumptions and biases. This is especially important when describing the discovery of significant or unanticipated findings.
V. Acknowledge the Study’s Limitations
It is far better for you to identify and acknowledge your study’s limitations than to have them pointed out by your professor! Note any unanswered questions or issues your study could not address and describe the generalizability of your results to other situations. If a limitation is applicable to the method chosen to gather information, then describe in detail the problems you encountered and why. VI. Make Suggestions for Further Research
You may choose to conclude the discussion section by making suggestions for further research [as opposed to offering suggestions in the conclusion of your paper]. Although your study can offer important insights about the research problem, this is where you can address other questions related to the problem that remain unanswered or highlight hidden issues that were revealed as a result of conducting your research. You should frame your suggestions by linking the need for further research to the limitations of your study [e.g., in future studies, the survey instrument should include more questions that ask..."] or linking to critical issues revealed from the data that were not considered initially in your research.
NOTE: Besides the literature review section, the preponderance of references to sources is usually found in the discussion section . A few historical references may be helpful for perspective, but most of the references should be relatively recent and included to aid in the interpretation of your results, to support the significance of a finding, and/or to place a finding within a particular context. If a study that you cited does not support your findings, don't ignore it--clearly explain why your research findings differ from theirs.
V. Problems to Avoid
- Do not waste time restating your results . Should you need to remind the reader of a finding to be discussed, use "bridge sentences" that relate the result to the interpretation. An example would be: “In the case of determining available housing to single women with children in rural areas of Texas, the findings suggest that access to good schools is important...," then move on to further explaining this finding and its implications.
- As noted, recommendations for further research can be included in either the discussion or conclusion of your paper, but do not repeat your recommendations in the both sections. Think about the overall narrative flow of your paper to determine where best to locate this information. However, if your findings raise a lot of new questions or issues, consider including suggestions for further research in the discussion section.
- Do not introduce new results in the discussion section. Be wary of mistaking the reiteration of a specific finding for an interpretation because it may confuse the reader. The description of findings [results section] and the interpretation of their significance [discussion section] should be distinct parts of your paper. If you choose to combine the results section and the discussion section into a single narrative, you must be clear in how you report the information discovered and your own interpretation of each finding. This approach is not recommended if you lack experience writing college-level research papers.
- Use of the first person pronoun is generally acceptable. Using first person singular pronouns can help emphasize a point or illustrate a contrasting finding. However, keep in mind that too much use of the first person can actually distract the reader from the main points [i.e., I know you're telling me this--just tell me!].
Analyzing vs. Summarizing. Department of English Writing Guide. George Mason University; Discussion. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College; Hess, Dean R. "How to Write an Effective Discussion." Respiratory Care 49 (October 2004); Kretchmer, Paul. Fourteen Steps to Writing to Writing an Effective Discussion Section. San Francisco Edit, 2003-2008; The Lab Report. University College Writing Centre. University of Toronto; Sauaia, A. et al. "The Anatomy of an Article: The Discussion Section: "How Does the Article I Read Today Change What I Will Recommend to my Patients Tomorrow?” The Journal of Trauma and Acute Care Surgery 74 (June 2013): 1599-1602; Research Limitations & Future Research . Lund Research Ltd., 2012; Summary: Using it Wisely. The Writing Center. University of North Carolina; Schafer, Mickey S. Writing the Discussion. Writing in Psychology course syllabus. University of Florida; Yellin, Linda L. A Sociology Writer's Guide . Boston, MA: Allyn and Bacon, 2009.
Writing Tip
Don’t Over-Interpret the Results!
Interpretation is a subjective exercise. As such, you should always approach the selection and interpretation of your findings introspectively and to think critically about the possibility of judgmental biases unintentionally entering into discussions about the significance of your work. With this in mind, be careful that you do not read more into the findings than can be supported by the evidence you have gathered. Remember that the data are the data: nothing more, nothing less.
MacCoun, Robert J. "Biases in the Interpretation and Use of Research Results." Annual Review of Psychology 49 (February 1998): 259-287; Ward, Paulet al, editors. The Oxford Handbook of Expertise . Oxford, UK: Oxford University Press, 2018.
Another Writing Tip
Don't Write Two Results Sections!
One of the most common mistakes that you can make when discussing the results of your study is to present a superficial interpretation of the findings that more or less re-states the results section of your paper. Obviously, you must refer to your results when discussing them, but focus on the interpretation of those results and their significance in relation to the research problem, not the data itself.
Azar, Beth. "Discussing Your Findings." American Psychological Association gradPSYCH Magazine (January 2006).
Yet Another Writing Tip
Avoid Unwarranted Speculation!
The discussion section should remain focused on the findings of your study. For example, if the purpose of your research was to measure the impact of foreign aid on increasing access to education among disadvantaged children in Bangladesh, it would not be appropriate to speculate about how your findings might apply to populations in other countries without drawing from existing studies to support your claim or if analysis of other countries was not a part of your original research design. If you feel compelled to speculate, do so in the form of describing possible implications or explaining possible impacts. Be certain that you clearly identify your comments as speculation or as a suggestion for where further research is needed. Sometimes your professor will encourage you to expand your discussion of the results in this way, while others don’t care what your opinion is beyond your effort to interpret the data in relation to the research problem.
- << Previous: Using Non-Textual Elements
- Next: Limitations of the Study >>
- Last Updated: Jun 18, 2024 10:45 AM
- URL: https://libguides.usc.edu/writingguide
- Affiliate Program

- UNITED STATES
- 台灣 (TAIWAN)
- TÜRKIYE (TURKEY)
- Academic Editing Services
- - Research Paper
- - Journal Manuscript
- - Dissertation
- - College & University Assignments
- Admissions Editing Services
- - Application Essay
- - Personal Statement
- - Recommendation Letter
- - Cover Letter
- - CV/Resume
- Business Editing Services
- - Business Documents
- - Report & Brochure
- - Website & Blog
- Writer Editing Services
- - Script & Screenplay
- Our Editors
- Client Reviews
- Editing & Proofreading Prices
- Wordvice Points
- Partner Discount
- Plagiarism Checker
- APA Citation Generator
- MLA Citation Generator
- Chicago Citation Generator
- Vancouver Citation Generator
- - APA Style
- - MLA Style
- - Chicago Style
- - Vancouver Style
- Writing & Editing Guide
- Academic Resources
- Admissions Resources
How to Write a Discussion Section for a Research Paper
We’ve talked about several useful writing tips that authors should consider while drafting or editing their research papers. In particular, we’ve focused on figures and legends , as well as the Introduction , Methods , and Results . Now that we’ve addressed the more technical portions of your journal manuscript, let’s turn to the analytical segments of your research article. In this article, we’ll provide tips on how to write a strong Discussion section that best portrays the significance of your research contributions.
What is the Discussion section of a research paper?
In a nutshell, your Discussion fulfills the promise you made to readers in your Introduction . At the beginning of your paper, you tell us why we should care about your research. You then guide us through a series of intricate images and graphs that capture all the relevant data you collected during your research. We may be dazzled and impressed at first, but none of that matters if you deliver an anti-climactic conclusion in the Discussion section!
Are you feeling pressured? Don’t worry. To be honest, you will edit the Discussion section of your manuscript numerous times. After all, in as little as one to two paragraphs ( Nature ‘s suggestion based on their 3,000-word main body text limit), you have to explain how your research moves us from point A (issues you raise in the Introduction) to point B (our new understanding of these matters). You must also recommend how we might get to point C (i.e., identify what you think is the next direction for research in this field). That’s a lot to say in two paragraphs!
So, how do you do that? Let’s take a closer look.
What should I include in the Discussion section?
As we stated above, the goal of your Discussion section is to answer the questions you raise in your Introduction by using the results you collected during your research . The content you include in the Discussions segment should include the following information:
- Remind us why we should be interested in this research project.
- Describe the nature of the knowledge gap you were trying to fill using the results of your study.
- Don’t repeat your Introduction. Instead, focus on why this particular study was needed to fill the gap you noticed and why that gap needed filling in the first place.
- Mainly, you want to remind us of how your research will increase our knowledge base and inspire others to conduct further research.
- Clearly tell us what that piece of missing knowledge was.
- Answer each of the questions you asked in your Introduction and explain how your results support those conclusions.
- Make sure to factor in all results relevant to the questions (even if those results were not statistically significant).
- Focus on the significance of the most noteworthy results.
- If conflicting inferences can be drawn from your results, evaluate the merits of all of them.
- Don’t rehash what you said earlier in the Results section. Rather, discuss your findings in the context of answering your hypothesis. Instead of making statements like “[The first result] was this…,” say, “[The first result] suggests [conclusion].”
- Do your conclusions line up with existing literature?
- Discuss whether your findings agree with current knowledge and expectations.
- Keep in mind good persuasive argument skills, such as explaining the strengths of your arguments and highlighting the weaknesses of contrary opinions.
- If you discovered something unexpected, offer reasons. If your conclusions aren’t aligned with current literature, explain.
- Address any limitations of your study and how relevant they are to interpreting your results and validating your findings.
- Make sure to acknowledge any weaknesses in your conclusions and suggest room for further research concerning that aspect of your analysis.
- Make sure your suggestions aren’t ones that should have been conducted during your research! Doing so might raise questions about your initial research design and protocols.
- Similarly, maintain a critical but unapologetic tone. You want to instill confidence in your readers that you have thoroughly examined your results and have objectively assessed them in a way that would benefit the scientific community’s desire to expand our knowledge base.
- Recommend next steps.
- Your suggestions should inspire other researchers to conduct follow-up studies to build upon the knowledge you have shared with them.
- Keep the list short (no more than two).
How to Write the Discussion Section
The above list of what to include in the Discussion section gives an overall idea of what you need to focus on throughout the section. Below are some tips and general suggestions about the technical aspects of writing and organization that you might find useful as you draft or revise the contents we’ve outlined above.
Technical writing elements
- Embrace active voice because it eliminates the awkward phrasing and wordiness that accompanies passive voice.
- Use the present tense, which should also be employed in the Introduction.
- Sprinkle with first person pronouns if needed, but generally, avoid it. We want to focus on your findings.
- Maintain an objective and analytical tone.
Discussion section organization
- Keep the same flow across the Results, Methods, and Discussion sections.
- We develop a rhythm as we read and parallel structures facilitate our comprehension. When you organize information the same way in each of these related parts of your journal manuscript, we can quickly see how a certain result was interpreted and quickly verify the particular methods used to produce that result.
- Notice how using parallel structure will eliminate extra narration in the Discussion part since we can anticipate the flow of your ideas based on what we read in the Results segment. Reducing wordiness is important when you only have a few paragraphs to devote to the Discussion section!
- Within each subpart of a Discussion, the information should flow as follows: (A) conclusion first, (B) relevant results and how they relate to that conclusion and (C) relevant literature.
- End with a concise summary explaining the big-picture impact of your study on our understanding of the subject matter. At the beginning of your Discussion section, you stated why this particular study was needed to fill the gap you noticed and why that gap needed filling in the first place. Now, it is time to end with “how your research filled that gap.”
Discussion Part 1: Summarizing Key Findings
Begin the Discussion section by restating your statement of the problem and briefly summarizing the major results. Do not simply repeat your findings. Rather, try to create a concise statement of the main results that directly answer the central research question that you stated in the Introduction section . This content should not be longer than one paragraph in length.
Many researchers struggle with understanding the precise differences between a Discussion section and a Results section . The most important thing to remember here is that your Discussion section should subjectively evaluate the findings presented in the Results section, and in relatively the same order. Keep these sections distinct by making sure that you do not repeat the findings without providing an interpretation.
Phrase examples: Summarizing the results
- The findings indicate that …
- These results suggest a correlation between A and B …
- The data present here suggest that …
- An interpretation of the findings reveals a connection between…
Discussion Part 2: Interpreting the Findings
What do the results mean? It may seem obvious to you, but simply looking at the figures in the Results section will not necessarily convey to readers the importance of the findings in answering your research questions.
The exact structure of interpretations depends on the type of research being conducted. Here are some common approaches to interpreting data:
- Identifying correlations and relationships in the findings
- Explaining whether the results confirm or undermine your research hypothesis
- Giving the findings context within the history of similar research studies
- Discussing unexpected results and analyzing their significance to your study or general research
- Offering alternative explanations and arguing for your position
Organize the Discussion section around key arguments, themes, hypotheses, or research questions or problems. Again, make sure to follow the same order as you did in the Results section.
Discussion Part 3: Discussing the Implications
In addition to providing your own interpretations, show how your results fit into the wider scholarly literature you surveyed in the literature review section. This section is called the implications of the study . Show where and how these results fit into existing knowledge, what additional insights they contribute, and any possible consequences that might arise from this knowledge, both in the specific research topic and in the wider scientific domain.
Questions to ask yourself when dealing with potential implications:
- Do your findings fall in line with existing theories, or do they challenge these theories or findings? What new information do they contribute to the literature, if any? How exactly do these findings impact or conflict with existing theories or models?
- What are the practical implications on actual subjects or demographics?
- What are the methodological implications for similar studies conducted either in the past or future?
Your purpose in giving the implications is to spell out exactly what your study has contributed and why researchers and other readers should be interested.
Phrase examples: Discussing the implications of the research
- These results confirm the existing evidence in X studies…
- The results are not in line with the foregoing theory that…
- This experiment provides new insights into the connection between…
- These findings present a more nuanced understanding of…
- While previous studies have focused on X, these results demonstrate that Y.
Step 4: Acknowledging the limitations
All research has study limitations of one sort or another. Acknowledging limitations in methodology or approach helps strengthen your credibility as a researcher. Study limitations are not simply a list of mistakes made in the study. Rather, limitations help provide a more detailed picture of what can or cannot be concluded from your findings. In essence, they help temper and qualify the study implications you listed previously.
Study limitations can relate to research design, specific methodological or material choices, or unexpected issues that emerged while you conducted the research. Mention only those limitations directly relate to your research questions, and explain what impact these limitations had on how your study was conducted and the validity of any interpretations.
Possible types of study limitations:
- Insufficient sample size for statistical measurements
- Lack of previous research studies on the topic
- Methods/instruments/techniques used to collect the data
- Limited access to data
- Time constraints in properly preparing and executing the study
After discussing the study limitations, you can also stress that your results are still valid. Give some specific reasons why the limitations do not necessarily handicap your study or narrow its scope.
Phrase examples: Limitations sentence beginners
- “There may be some possible limitations in this study.”
- “The findings of this study have to be seen in light of some limitations.”
- “The first limitation is the…The second limitation concerns the…”
- “The empirical results reported herein should be considered in the light of some limitations.”
- “This research, however, is subject to several limitations.”
- “The primary limitation to the generalization of these results is…”
- “Nonetheless, these results must be interpreted with caution and a number of limitations should be borne in mind.”
Discussion Part 5: Giving Recommendations for Further Research
Based on your interpretation and discussion of the findings, your recommendations can include practical changes to the study or specific further research to be conducted to clarify the research questions. Recommendations are often listed in a separate Conclusion section , but often this is just the final paragraph of the Discussion section.
Suggestions for further research often stem directly from the limitations outlined. Rather than simply stating that “further research should be conducted,” provide concrete specifics for how future can help answer questions that your research could not.
Phrase examples: Recommendation sentence beginners
- Further research is needed to establish …
- There is abundant space for further progress in analyzing…
- A further study with more focus on X should be done to investigate…
- Further studies of X that account for these variables must be undertaken.
Consider Receiving Professional Language Editing
As you edit or draft your research manuscript, we hope that you implement these guidelines to produce a more effective Discussion section. And after completing your draft, don’t forget to submit your work to a professional proofreading and English editing service like Wordvice, including our manuscript editing service for paper editing , cover letter editing , SOP editing , and personal statement proofreading services. Language editors not only proofread and correct errors in grammar, punctuation, mechanics, and formatting but also improve terms and revise phrases so they read more naturally. Wordvice is an industry leader in providing high-quality revision for all types of academic documents.
For additional information about how to write a strong research paper, make sure to check out our full research writing series !
Wordvice Writing Resources
- How to Write a Research Paper Introduction
- Which Verb Tenses to Use in a Research Paper
- How to Write an Abstract for a Research Paper
- How to Write a Research Paper Title
- Useful Phrases for Academic Writing
- Common Transition Terms in Academic Papers
- Active and Passive Voice in Research Papers
- 100+ Verbs That Will Make Your Research Writing Amazing
- Tips for Paraphrasing in Research Papers
Additional Academic Resources
- Guide for Authors. (Elsevier)
- How to Write the Results Section of a Research Paper. (Bates College)
- Structure of a Research Paper. (University of Minnesota Biomedical Library)
- How to Choose a Target Journal (Springer)
- How to Write Figures and Tables (UNC Writing Center)

- Langson Library
- Science Library
- Grunigen Medical Library
- Law Library
- Connect From Off-Campus
- Accessibility
- Gateway Study Center

Email this link
Writing a scientific paper.
- Writing a lab report
- INTRODUCTION
Writing a "good" discussion section
"discussion and conclusions checklist" from: how to write a good scientific paper. chris a. mack. spie. 2018., peer review.
- LITERATURE CITED
- Bibliography of guides to scientific writing and presenting
- Presentations
- Lab Report Writing Guides on the Web
This is is usually the hardest section to write. You are trying to bring out the true meaning of your data without being too long. Do not use words to conceal your facts or reasoning. Also do not repeat your results, this is a discussion.
- Present principles, relationships and generalizations shown by the results
- Point out exceptions or lack of correlations. Define why you think this is so.
- Show how your results agree or disagree with previously published works
- Discuss the theoretical implications of your work as well as practical applications
- State your conclusions clearly. Summarize your evidence for each conclusion.
- Discuss the significance of the results
- Evidence does not explain itself; the results must be presented and then explained.
- Typical stages in the discussion: summarizing the results, discussing whether results are expected or unexpected, comparing these results to previous work, interpreting and explaining the results (often by comparison to a theory or model), and hypothesizing about their generality.
- Discuss any problems or shortcomings encountered during the course of the work.
- Discuss possible alternate explanations for the results.
- Avoid: presenting results that are never discussed; presenting discussion that does not relate to any of the results; presenting results and discussion in chronological order rather than logical order; ignoring results that do not support the conclusions; drawing conclusions from results without logical arguments to back them up.
CONCLUSIONS
- Provide a very brief summary of the Results and Discussion.
- Emphasize the implications of the findings, explaining how the work is significant and providing the key message(s) the author wishes to convey.
- Provide the most general claims that can be supported by the evidence.
- Provide a future perspective on the work.
- Avoid: repeating the abstract; repeating background information from the Introduction; introducing new evidence or new arguments not found in the Results and Discussion; repeating the arguments made in the Results and Discussion; failing to address all of the research questions set out in the Introduction.
WHAT HAPPENS AFTER I COMPLETE MY PAPER?
The peer review process is the quality control step in the publication of ideas. Papers that are submitted to a journal for publication are sent out to several scientists (peers) who look carefully at the paper to see if it is "good science". These reviewers then recommend to the editor of a journal whether or not a paper should be published. Most journals have publication guidelines. Ask for them and follow them exactly. Peer reviewers examine the soundness of the materials and methods section. Are the materials and methods used written clearly enough for another scientist to reproduce the experiment? Other areas they look at are: originality of research, significance of research question studied, soundness of the discussion and interpretation, correct spelling and use of technical terms, and length of the article.
- << Previous: RESULTS
- Next: LITERATURE CITED >>
- Last Updated: Aug 4, 2023 9:33 AM
- URL: https://guides.lib.uci.edu/scientificwriting
Off-campus? Please use the Software VPN and choose the group UCIFull to access licensed content. For more information, please Click here
Software VPN is not available for guests, so they may not have access to some content when connecting from off-campus.
Guide to Writing the Results and Discussion Sections of a Scientific Article
A quality research paper has both the qualities of in-depth research and good writing ( Bordage, 2001 ). In addition, a research paper must be clear, concise, and effective when presenting the information in an organized structure with a logical manner ( Sandercock, 2013 ).
In this article, we will take a closer look at the results and discussion section. Composing each of these carefully with sufficient data and well-constructed arguments can help improve your paper overall.

The results section of your research paper contains a description about the main findings of your research, whereas the discussion section interprets the results for readers and provides the significance of the findings. The discussion should not repeat the results.
Let’s dive in a little deeper about how to properly, and clearly organize each part.
How to Organize the Results Section
Since your results follow your methods, you’ll want to provide information about what you discovered from the methods you used, such as your research data. In other words, what were the outcomes of the methods you used?
You may also include information about the measurement of your data, variables, treatments, and statistical analyses.
To start, organize your research data based on how important those are in relation to your research questions. This section should focus on showing major results that support or reject your research hypothesis. Include your least important data as supplemental materials when submitting to the journal.
The next step is to prioritize your research data based on importance – focusing heavily on the information that directly relates to your research questions using the subheadings.
The organization of the subheadings for the results section usually mirrors the methods section. It should follow a logical and chronological order.
Subheading organization
Subheadings within your results section are primarily going to detail major findings within each important experiment. And the first paragraph of your results section should be dedicated to your main findings (findings that answer your overall research question and lead to your conclusion) (Hofmann, 2013).
In the book “Writing in the Biological Sciences,” author Angelika Hofmann recommends you structure your results subsection paragraphs as follows:
- Experimental purpose
- Interpretation
Each subheading may contain a combination of ( Bahadoran, 2019 ; Hofmann, 2013, pg. 62-63):
- Text: to explain about the research data
- Figures: to display the research data and to show trends or relationships, for examples using graphs or gel pictures.
- Tables: to represent a large data and exact value
Decide on the best way to present your data — in the form of text, figures or tables (Hofmann, 2013).
Data or Results?
Sometimes we get confused about how to differentiate between data and results . Data are information (facts or numbers) that you collected from your research ( Bahadoran, 2019 ).

Whereas, results are the texts presenting the meaning of your research data ( Bahadoran, 2019 ).

One mistake that some authors often make is to use text to direct the reader to find a specific table or figure without further explanation. This can confuse readers when they interpret data completely different from what the authors had in mind. So, you should briefly explain your data to make your information clear for the readers.
Common Elements in Figures and Tables
Figures and tables present information about your research data visually. The use of these visual elements is necessary so readers can summarize, compare, and interpret large data at a glance. You can use graphs or figures to compare groups or patterns. Whereas, tables are ideal to present large quantities of data and exact values.
Several components are needed to create your figures and tables. These elements are important to sort your data based on groups (or treatments). It will be easier for the readers to see the similarities and differences among the groups.
When presenting your research data in the form of figures and tables, organize your data based on the steps of the research leading you into a conclusion.
Common elements of the figures (Bahadoran, 2019):
- Figure number
- Figure title
- Figure legend (for example a brief title, experimental/statistical information, or definition of symbols).
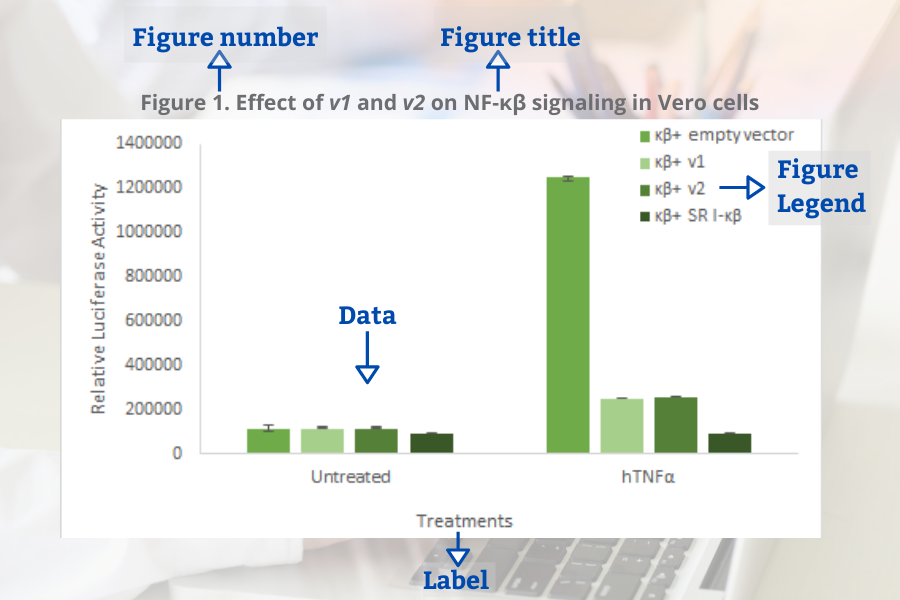
Tables in the result section may contain several elements (Bahadoran, 2019):
- Table number
- Table title
- Row headings (for example groups)
- Column headings
- Row subheadings (for example categories or groups)
- Column subheadings (for example categories or variables)
- Footnotes (for example statistical analyses)
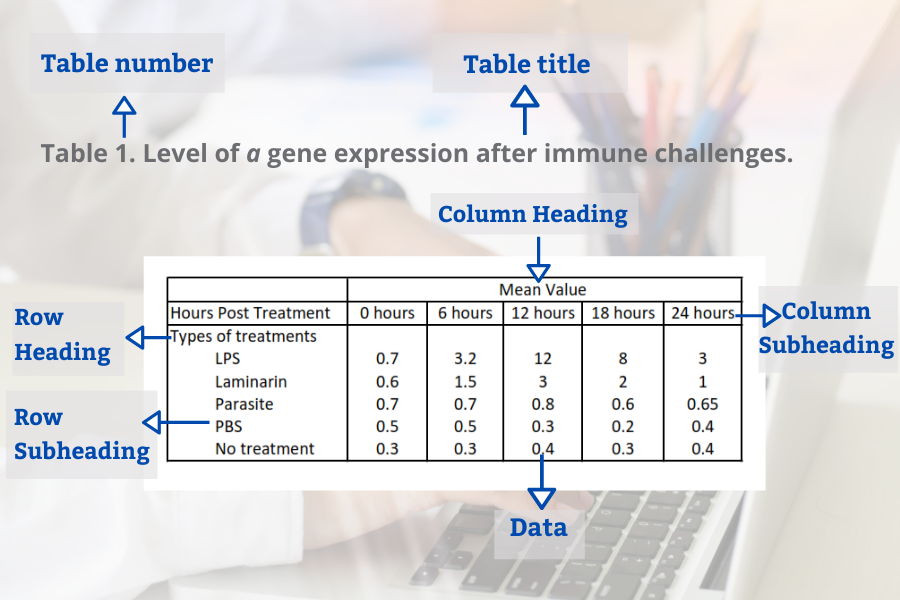
Tips to Write the Results Section
- Direct the reader to the research data and explain the meaning of the data.
- Avoid using a repetitive sentence structure to explain a new set of data.
- Write and highlight important findings in your results.
- Use the same order as the subheadings of the methods section.
- Match the results with the research questions from the introduction. Your results should answer your research questions.
- Be sure to mention the figures and tables in the body of your text.
- Make sure there is no mismatch between the table number or the figure number in text and in figure/tables.
- Only present data that support the significance of your study. You can provide additional data in tables and figures as supplementary material.
How to Organize the Discussion Section
It’s not enough to use figures and tables in your results section to convince your readers about the importance of your findings. You need to support your results section by providing more explanation in the discussion section about what you found.
In the discussion section, based on your findings, you defend the answers to your research questions and create arguments to support your conclusions.
Below is a list of questions to guide you when organizing the structure of your discussion section ( Viera et al ., 2018 ):
- What experiments did you conduct and what were the results?
- What do the results mean?
- What were the important results from your study?
- How did the results answer your research questions?
- Did your results support your hypothesis or reject your hypothesis?
- What are the variables or factors that might affect your results?
- What were the strengths and limitations of your study?
- What other published works support your findings?
- What other published works contradict your findings?
- What possible factors might cause your findings different from other findings?
- What is the significance of your research?
- What are new research questions to explore based on your findings?
Organizing the Discussion Section
The structure of the discussion section may be different from one paper to another, but it commonly has a beginning, middle-, and end- to the section.
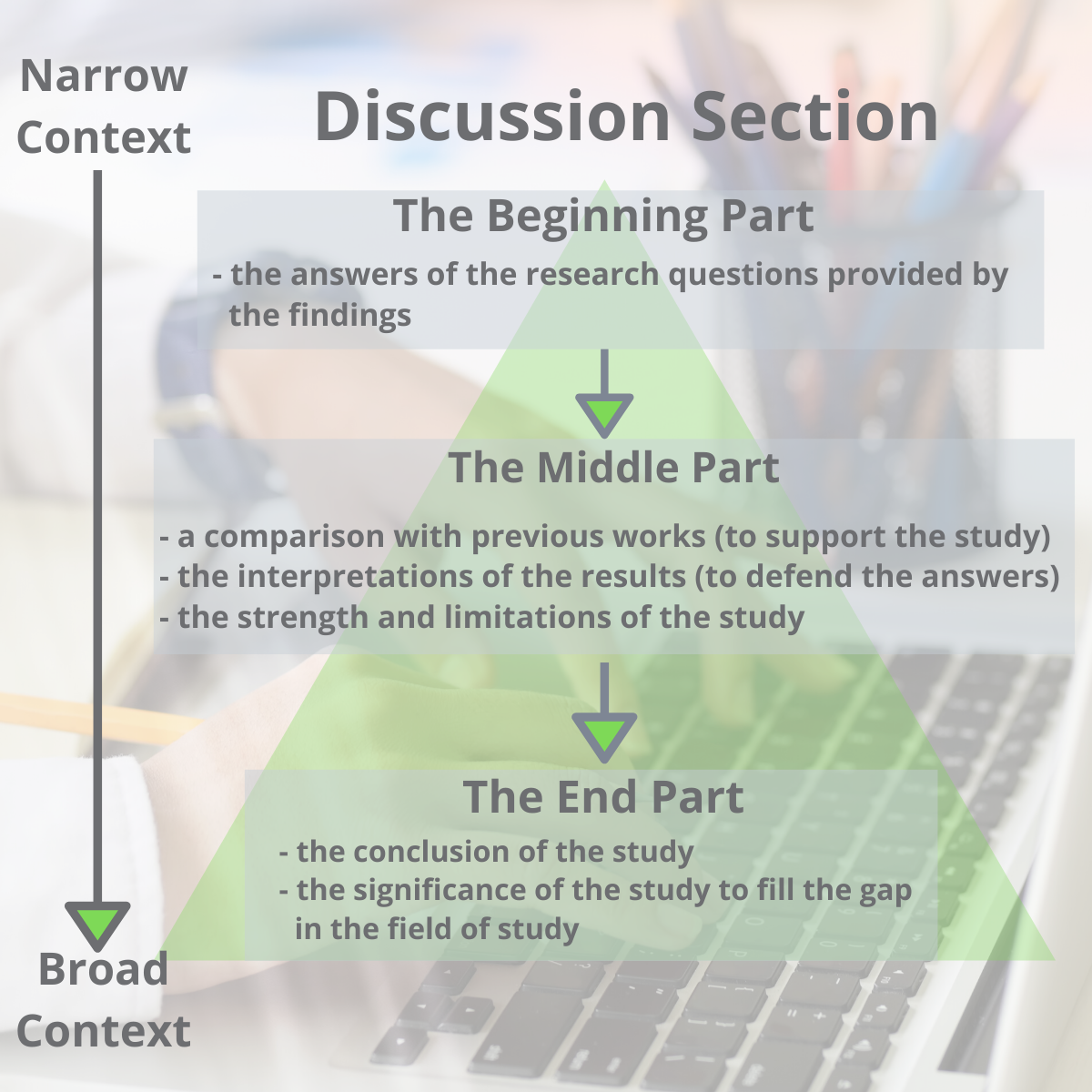
One way to organize the structure of the discussion section is by dividing it into three parts (Ghasemi, 2019):
- The beginning: The first sentence of the first paragraph should state the importance and the new findings of your research. The first paragraph may also include answers to your research questions mentioned in your introduction section.
- The middle: The middle should contain the interpretations of the results to defend your answers, the strength of the study, the limitations of the study, and an update literature review that validates your findings.
- The end: The end concludes the study and the significance of your research.
Another possible way to organize the discussion section was proposed by Michael Docherty in British Medical Journal: is by using this structure ( Docherty, 1999 ):
- Discussion of important findings
- Comparison of your results with other published works
- Include the strengths and limitations of the study
- Conclusion and possible implications of your study, including the significance of your study – address why and how is it meaningful
- Future research questions based on your findings
Finally, a last option is structuring your discussion this way (Hofmann, 2013, pg. 104):
- First Paragraph: Provide an interpretation based on your key findings. Then support your interpretation with evidence.
- Secondary results
- Limitations
- Unexpected findings
- Comparisons to previous publications
- Last Paragraph: The last paragraph should provide a summarization (conclusion) along with detailing the significance, implications and potential next steps.
Remember, at the heart of the discussion section is presenting an interpretation of your major findings.
Tips to Write the Discussion Section
- Highlight the significance of your findings
- Mention how the study will fill a gap in knowledge.
- Indicate the implication of your research.
- Avoid generalizing, misinterpreting your results, drawing a conclusion with no supportive findings from your results.
Aggarwal, R., & Sahni, P. (2018). The Results Section. In Reporting and Publishing Research in the Biomedical Sciences (pp. 21-38): Springer.
Bahadoran, Z., Mirmiran, P., Zadeh-Vakili, A., Hosseinpanah, F., & Ghasemi, A. (2019). The principles of biomedical scientific writing: Results. International journal of endocrinology and metabolism, 17(2).
Bordage, G. (2001). Reasons reviewers reject and accept manuscripts: the strengths and weaknesses in medical education reports. Academic medicine, 76(9), 889-896.
Cals, J. W., & Kotz, D. (2013). Effective writing and publishing scientific papers, part VI: discussion. Journal of clinical epidemiology, 66(10), 1064.
Docherty, M., & Smith, R. (1999). The case for structuring the discussion of scientific papers: Much the same as that for structuring abstracts. In: British Medical Journal Publishing Group.
Faber, J. (2017). Writing scientific manuscripts: most common mistakes. Dental press journal of orthodontics, 22(5), 113-117.
Fletcher, R. H., & Fletcher, S. W. (2018). The discussion section. In Reporting and Publishing Research in the Biomedical Sciences (pp. 39-48): Springer.
Ghasemi, A., Bahadoran, Z., Mirmiran, P., Hosseinpanah, F., Shiva, N., & Zadeh-Vakili, A. (2019). The Principles of Biomedical Scientific Writing: Discussion. International journal of endocrinology and metabolism, 17(3).
Hofmann, A. H. (2013). Writing in the biological sciences: a comprehensive resource for scientific communication . New York: Oxford University Press.
Kotz, D., & Cals, J. W. (2013). Effective writing and publishing scientific papers, part V: results. Journal of clinical epidemiology, 66(9), 945.
Mack, C. (2014). How to Write a Good Scientific Paper: Structure and Organization. Journal of Micro/ Nanolithography, MEMS, and MOEMS, 13. doi:10.1117/1.JMM.13.4.040101
Moore, A. (2016). What's in a Discussion section? Exploiting 2‐dimensionality in the online world…. Bioessays, 38(12), 1185-1185.
Peat, J., Elliott, E., Baur, L., & Keena, V. (2013). Scientific writing: easy when you know how: John Wiley & Sons.
Sandercock, P. M. L. (2012). How to write and publish a scientific article. Canadian Society of Forensic Science Journal, 45(1), 1-5.
Teo, E. K. (2016). Effective Medical Writing: The Write Way to Get Published. Singapore Medical Journal, 57(9), 523-523. doi:10.11622/smedj.2016156
Van Way III, C. W. (2007). Writing a scientific paper. Nutrition in Clinical Practice, 22(6), 636-640.
Vieira, R. F., Lima, R. C. d., & Mizubuti, E. S. G. (2019). How to write the discussion section of a scientific article. Acta Scientiarum. Agronomy, 41.
Related Articles

A quality research paper has both the qualities of in-depth research and good writing (Bordage, 200...

How to Survive and Complete a Thesis or a Dissertation
Writing a thesis or a dissertation can be a challenging process for many graduate students. There ar...

12 Ways to Dramatically Improve your Research Manuscript Title and Abstract
The first thing a person doing literary research will see is a research publication title. After tha...

15 Laboratory Notebook Tips to Help with your Research Manuscript
Your lab notebook is a foundation to your research manuscript. It serves almost as a rudimentary dra...
Join our list to receive promos and articles.

- Competent Cells
- Lab Startup
- Z')" data-type="collection" title="Products A->Z" target="_self" href="/collection/products-a-to-z">Products A->Z
- GoldBio Resources
- GoldBio Sales Team
- GoldBio Distributors
- Duchefa Direct
- Sign up for Promos
- Terms & Conditions
- ISO Certification
- Agarose Resins
- Antibiotics & Selection
- Biochemical Reagents
- Bioluminescence
- Buffers & Reagents
- Cell Culture
- Cloning & Induction
- Competent Cells and Transformation
- Detergents & Membrane Agents
- DNA Amplification
- Enzymes, Inhibitors & Substrates
- Growth Factors and Cytokines
- Lab Tools & Accessories
- Plant Research and Reagents
- Protein Research & Analysis
- Protein Expression & Purification
- Reducing Agents

- Walden University
- Faculty Portal
General Research Paper Guidelines: Discussion
Discussion section.
The overall purpose of a research paper’s discussion section is to evaluate and interpret results, while explaining both the implications and limitations of your findings. Per APA (2020) guidelines, this section requires you to “examine, interpret, and qualify the results and draw inferences and conclusions from them” (p. 89). Discussion sections also require you to detail any new insights, think through areas for future research, highlight the work that still needs to be done to further your topic, and provide a clear conclusion to your research paper. In a good discussion section, you should do the following:
- Clearly connect the discussion of your results to your introduction, including your central argument, thesis, or problem statement.
- Provide readers with a critical thinking through of your results, answering the “so what?” question about each of your findings. In other words, why is this finding important?
- Detail how your research findings might address critical gaps or problems in your field
- Compare your results to similar studies’ findings
- Provide the possibility of alternative interpretations, as your goal as a researcher is to “discover” and “examine” and not to “prove” or “disprove.” Instead of trying to fit your results into your hypothesis, critically engage with alternative interpretations to your results.
For more specific details on your Discussion section, be sure to review Sections 3.8 (pp. 89-90) and 3.16 (pp. 103-104) of your 7 th edition APA manual
*Box content adapted from:
University of Southern California (n.d.). Organizing your social sciences research paper: 8 the discussion . https://libguides.usc.edu/writingguide/discussion
Limitations
Limitations of generalizability or utility of findings, often over which the researcher has no control, should be detailed in your Discussion section. Including limitations for your reader allows you to demonstrate you have thought critically about your given topic, understood relevant literature addressing your topic, and chosen the methodology most appropriate for your research. It also allows you an opportunity to suggest avenues for future research on your topic. An effective limitations section will include the following:
- Detail (a) sources of potential bias, (b) possible imprecision of measures, (c) other limitations or weaknesses of the study, including any methodological or researcher limitations.
- Sample size: In quantitative research, if a sample size is too small, it is more difficult to generalize results.
- Lack of available/reliable data : In some cases, data might not be available or reliable, which will ultimately affect the overall scope of your research. Use this as an opportunity to explain areas for future study.
- Lack of prior research on your study topic: In some cases, you might find that there is very little or no similar research on your study topic, which hinders the credibility and scope of your own research. If this is the case, use this limitation as an opportunity to call for future research. However, make sure you have done a thorough search of the available literature before making this claim.
- Flaws in measurement of data: Hindsight is 20/20, and you might realize after you have completed your research that the data tool you used actually limited the scope or results of your study in some way. Again, acknowledge the weakness and use it as an opportunity to highlight areas for future study.
- Limits of self-reported data: In your research, you are assuming that any participants will be honest and forthcoming with responses or information they provide to you. Simply acknowledging this assumption as a possible limitation is important in your research.
- Access: Most research requires that you have access to people, documents, organizations, etc.. However, for various reasons, access is sometimes limited or denied altogether. If this is the case, you will want to acknowledge access as a limitation to your research.
- Time: Choosing a research focus that is narrow enough in scope to finish in a given time period is important. If such limitations of time prevent you from certain forms of research, access, or study designs, acknowledging this time restraint is important. Acknowledging such limitations is important, as they can point other researchers to areas that require future study.
- Potential Bias: All researchers have some biases, so when reading and revising your draft, pay special attention to the possibilities for bias in your own work. Such bias could be in the form you organized people, places, participants, or events. They might also exist in the method you selected or the interpretation of your results. Acknowledging such bias is an important part of the research process.
- Language Fluency: On occasion, researchers or research participants might have language fluency issues, which could potentially hinder results or how effectively you interpret results. If this is an issue in your research, make sure to acknowledge it in your limitations section.
University of Southern California (n.d.). Organizing your social sciences research paper: Limitations of the study . https://libguides.usc.edu/writingguide/limitations
In many research papers, the conclusion, like the limitations section, is folded into the larger discussion section. If you are unsure whether to include the conclusion as part of your discussion or as a separate section, be sure to defer to the assignment instructions or ask your instructor.
The conclusion is important, as it is specifically designed to highlight your research’s larger importance outside of the specific results of your study. Your conclusion section allows you to reiterate the main findings of your study, highlight their importance, and point out areas for future research. Based on the scope of your paper, your conclusion could be anywhere from one to three paragraphs long. An effective conclusion section should include the following:
- Describe the possibilities for continued research on your topic, including what might be improved, adapted, or added to ensure useful and informed future research.
- Provide a detailed account of the importance of your findings
- Reiterate why your problem is important, detail how your interpretation of results impacts the subfield of study, and what larger issues both within and outside of your field might be affected from such results
University of Southern California (n.d.). Organizing your social sciences research paper: 9. the conclusion . https://libguides.usc.edu/writingguide/conclusion
- Previous Page: Results
- Next Page: References
- Office of Student Disability Services
Walden Resources
Departments.
- Academic Residencies
- Academic Skills
- Career Planning and Development
- Customer Care Team
- Field Experience
- Military Services
- Student Success Advising
- Writing Skills
Centers and Offices
- Center for Social Change
- Office of Academic Support and Instructional Services
- Office of Degree Acceleration
- Office of Research and Doctoral Services
- Office of Student Affairs
Student Resources
- Doctoral Writing Assessment
- Form & Style Review
- Quick Answers
- ScholarWorks
- SKIL Courses and Workshops
- Walden Bookstore
- Walden Catalog & Student Handbook
- Student Safety/Title IX
- Legal & Consumer Information
- Website Terms and Conditions
- Cookie Policy
- Accessibility
- Accreditation
- State Authorization
- Net Price Calculator
- Contact Walden
Walden University is a member of Adtalem Global Education, Inc. www.adtalem.com Walden University is certified to operate by SCHEV © 2024 Walden University LLC. All rights reserved.
- Our Writers
- How to Order
- Assignment Writing Service
- Report Writing Service
- Buy Coursework
- Dissertation Writing Service
- Research Paper Writing Service
- All Essay Services
Research Paper Guide
Research Paper Discussion Section
How To Write A Discussion For A Research Paper | Examples & Tips

People also read
Research Paper Writing - A Step by Step Guide
Research Paper Examples - Free Sample Papers for Different Formats!
Guide to Creating Effective Research Paper Outline
Interesting Research Paper Topics for 2024
Research Proposal Writing - A Step-by-Step Guide
How to Start a Research Paper - 7 Easy Steps
How to Write an Abstract for a Research Paper - A Step by Step Guide
Writing a Literature Review For a Research Paper - A Comprehensive Guide
Qualitative Research - Methods, Types, and Examples
8 Types of Qualitative Research - Overview & Examples
Qualitative vs Quantitative Research - Learning the Basics
200+ Engaging Psychology Research Paper Topics for Students in 2024
Learn How to Write a Hypothesis in a Research Paper: Examples and Tips!
20+ Types of Research With Examples - A Detailed Guide
Understanding Quantitative Research - Types & Data Collection Techniques
230+ Sociology Research Topics & Ideas for Students
How to Cite a Research Paper - A Complete Guide
Excellent History Research Paper Topics- 300+ Ideas
A Guide on Writing the Method Section of a Research Paper - Examples & Tips
How To Write an Introduction Paragraph For a Research Paper: Learn with Examples
Crafting a Winning Research Paper Title: A Complete Guide
Writing a Research Paper Conclusion - Step-by-Step Guide
Writing a Thesis For a Research Paper - A Comprehensive Guide
How To Write The Results Section of A Research Paper | Steps & Examples
Writing a Problem Statement for a Research Paper - A Comprehensive Guide
Finding Sources For a Research Paper: A Complete Guide
A Guide on How to Edit a Research Paper
200+ Ethical Research Paper Topics to Begin With (2024)
300+ Controversial Research Paper Topics & Ideas - 2024 Edition
150+ Argumentative Research Paper Topics For You - 2024
How to Write a Research Methodology for a Research Paper
Ever find yourself stuck when trying to write the discussion part of your research paper? Don't worry, it happens to a lot of people.
The discussion section is super important in your research paper . It's where you explain what your results mean. But turning all that data into a clear and meaningful story? That's not easy.
Guess what? MyPerfectWords.com has come up with a solution.
This blog is your guide to writing an outstanding discussion section. We'll guide you step by step with useful tips to make sure your research stands out.
So, let’s get started!
- 1. What Exactly is a Discussion Section in the Research Paper?
- 2. How to Write the Discussion Section of a Research Paper?
- 3. Examples of Good Discussion for a Research Paper
- 4. Mistakes to Avoid in Your Research Paper's Discussion
What Exactly is a Discussion Section in the Research Paper?
In a research paper, the discussion section is where you explain what your results really mean. It's like answering the questions, "So what?" and "What's the big picture?"
The discussion section is your chance to help your readers understand why your findings are important and how they fit into the larger context. It's more than just summarizing; it's about making your research understandable and meaningful to others.
Importance of the Discussion Section
The discussion section isn't just a formality; it's the heart of your research paper. This is where your findings transform from data into knowledge.
Let's break down why it's so crucial:
- Interpretation of Results : The discussion is where you get to tell readers what your results really mean. You go into the details, helping them understand the story behind the numbers or findings.
- Connecting the Dots : You connect different parts of your research, showing how they relate. This helps your readers see the bigger picture.
- Relevance to the Big Picture : You get to highlight why your research matters. How does it contribute to the broader understanding of the topic? This is your time to make your research significant.
- Addressing Limitations : In the discussion, you can acknowledge any limitations in your study and discuss how they might impact your results.
- Suggestions for Further Research : The discussion is where you suggest areas for future exploration. It's like passing the baton to the next researcher, indicating where more work could be done.

Tough Essay Due? Hire Tough Writers!
How to Write the Discussion Section of a Research Paper?
The Discussion section in a research paper plays a vital role in interpreting findings and formulating a conclusion . Given below are the main components of the discussion section:
- Quick Summary: A brief recap of your main findings.
- Interpretation: Significance and meaning of your results in relation to your research question.
- Literature Review : Connecting your findings with previous research or similar studies.
- Limitations: Discussing any study limitations, addressing potential concerns.
- Implications: Broader implications of your findings, considering practical and theoretical aspects.
- Alternative Explanations: Evaluating alternative interpretations, demonstrating a comprehensive analysis.
- Connecting to Hypotheses : Summarizing how your result section aligns or diverges from your initial hypotheses.
Now let’s explore the steps to write an effective discussion section that will effectively communicate the significance of your research:
Step 1: Get Started with a Quick Summary
Start by quickly telling your readers the main things you found in your research. Don't explain them in detail just yet; just give a simple overview.
This helps your readers get the big picture before diving into the details.
For instance, you conducted a study on the effects of exercise on mood. Your concise summary might look like this:
|
Step 2: Interpret Your Results
In the next step, talk about what your findings really mean. Share why the information you gathered is important. Connect each result to the questions you were trying to answer and the goals you set for your research.
You did research on whether plants grow better with different types of light. Here's how you interpret the results:
|
Step 3: Relate to Existing Literature
In this step, link up your discoveries with what other researchers have already figured out.
Share if your results are similar to or different from what's been found before. This helps give more background to your study and shows you know what other scientists have been up to.
You conducted a study on the impact of technology use on sleep patterns. Here’s how you can relate it to the existing knowledge or research:
|
Step 4: Address Limitations Honestly
Every study has its limitations. Acknowledge them openly in your discussion. This not only shows transparency but also helps readers interpret your results more accurately.
Let's consider a study on the effects of a new teaching method on student performance. You can address the limitations of the research like this: " |
Step 5: Discuss the Implications
Explore the implications of your findings. How do they contribute to the field? What real-world applications or changes might they suggest?
Dig into why your discoveries are important. How do they help the subject you studied?
This step is like looking at the bigger picture and asking, "So, what can we do with this information?"
Consider the example of a study on the impact of a new app on improving language learning; here’s how you can discuss your implications:
|
Paper Due? Why Suffer? That's our Job!
Step 6: Consider Alternative Explanations
After discussing the implications, challenge yourself by exploring alternative explanations for your results.
Discuss different perspectives and show that you've considered multiple angles.
Consider the example of a study on the impact of a new study technique on exam performance. This is how you consider alternative explanations:
|
Step 7: Connect to Your Hypotheses or Research Questions
For the last step, revisit your initial hypotheses or research questions. Explain whether your results support what you thought might happen or if they surprised you.
For instance, in a study about the impact of a new teaching method on student engagement, you can connect hypotheses like this:
|
Examples of Good Discussion for a Research Paper
Learning from well-crafted discussions can significantly enhance your own writing. Given below are some examples to help you understand how to write your own.
Discussion for a Research Paper Example Pdf
Discussion for a Medical Research Paper
Discussion Section for a Qualitative Research Paper
Mistakes to Avoid in Your Research Paper's Discussion
Writing the discussion section of your research paper can be tricky. To make sure you're on the right track, be mindful of these common mistakes:
- Overstating or Overinterpreting Results
Avoid making your findings sound more groundbreaking than they are. Stick to what your data actually shows, and don't exaggerate.
- Neglecting Alternative Explanations
Failing to consider other possible explanations for your results can weaken your discussion. Always explore alternative perspectives to present a well-rounded view.
- Ignoring Limitations
Don't sweep limitations under the rug. Acknowledge them openly and discuss how they might affect the validity or generalizability of your results.
- Being Overly Technical or Jargon-laden
Remember that your audience may not be experts in your specific field. Avoid using overly technical language or excessive jargon that could alienate your readers.
- Disregarding the 'So What' Factor
Always explain the significance of your findings. Don't leave your readers wondering why your research matters or how it contributes to the broader understanding of the subject.
- Rushing the Conclusion
The conclusion section of your discussion is critical. Don't rush it. Summarize the key points and leave your readers with a strong understanding of the significance of your research.
So, there you have it —writing a discussion and conclusion section isn't easy, but avoiding some common mistakes can make it much smoother.
Remember to keep it real with your results, think about what else could explain things, and don't forget about any limits in your study.
But if you're feeling stuck, MyPerfectWords.com is here for you.
Our team of experts knows their way around discussions. Whether you need some guidance or want someone to handle the writing for you, we've got your back.
Don't let discussion writing stress you out. Let our essay writing service for college make your academic life easier.

Write Essay Within 60 Seconds!

Dr. Barbara is a highly experienced writer and author who holds a Ph.D. degree in public health from an Ivy League school. She has worked in the medical field for many years, conducting extensive research on various health topics. Her writing has been featured in several top-tier publications.

Paper Due? Why Suffer? That’s our Job!
Keep reading
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Turk J Urol
- v.39(Suppl 1); 2013 Sep
How to write a discussion section?
Writing manuscripts to describe study outcomes, although not easy, is the main task of an academician. The aim of the present review is to outline the main aspects of writing the discussion section of a manuscript. Additionally, we address various issues regarding manuscripts in general. It is advisable to work on a manuscript regularly to avoid losing familiarity with the article. On principle, simple, clear and effective language should be used throughout the text. In addition, a pre-peer review process is recommended to obtain feedback on the manuscript. The discussion section can be written in 3 parts: an introductory paragraph, intermediate paragraphs and a conclusion paragraph. For intermediate paragraphs, a “divide and conquer” approach, meaning a full paragraph describing each of the study endpoints, can be used. In conclusion, academic writing is similar to other skills, and practice makes perfect.
Introduction
Sharing knowledge produced during academic life is achieved through writing manuscripts. However writing manuscripts is a challenging endeavour in that we physicians have a heavy workload, and English which is common language used for the dissemination of scientific knowledge is not our mother tongue.
The objective of this review is to summarize the method of writing ‘Discussion’ section which is the most important, but probably at the same time the most unlikable part of a manuscript, and demonstrate the easy ways we applied in our practice, and finally share the frequently made relevant mistakes. During this procedure, inevitably some issues which concerns general concept of manuscript writing process are dealt with. Therefore in this review we will deal with topics related to the general aspects of manuscript writing process, and specifically issues concerning only the ‘Discussion’ section.
A) Approaches to general aspects of manuscript writing process:
1. what should be the strategy of sparing time for manuscript writing be.
Two different approaches can be formulated on this issue? One of them is to allocate at least 30 minutes a day for writing a manuscript which amounts to 3.5 hours a week. This period of time is adequate for completion of a manuscript within a few weeks which can be generally considered as a long time interval. Fundamental advantage of this approach is to gain a habit of making academic researches if one complies with the designated time schedule, and to keep the manuscript writing motivation at persistently high levels. Another approach concerning this issue is to accomplish manuscript writing process within a week. With the latter approach, the target is rapidly attained. However longer time periods spent in order to concentrate on the subject matter can be boring, and lead to loss of motivation. Daily working requirements unrelated to the manuscript writing might intervene, and prolong manuscript writing process. Alienation periods can cause loss of time because of need for recurrent literature reviews. The most optimal approach to manuscript writing process is daily writing strategy where higher levels of motivation are persistently maintained.
Especially before writing the manuscript, the most important step at the start is to construct a draft, and completion of the manuscript on a theoretical basis. Therefore, during construction of a draft, attention distracting environment should be avoided, and this step should be completed within 1–2 hours. On the other hand, manuscript writing process should begin before the completion of the study (even the during project stage). The justification of this approach is to see the missing aspects of the study and the manuscript writing methodology, and try to solve the relevant problems before completion of the study. Generally, after completion of the study, it is very difficult to solve the problems which might be discerned during the writing process. Herein, at least drafts of the ‘Introduction’, and ‘Material and Methods’ can be written, and even tables containing numerical data can be constructed. These tables can be written down in the ‘Results’ section. [ 1 ]
2. How should the manuscript be written?
The most important principle to be remembered on this issue is to obey the criteria of simplicity, clarity, and effectiveness. [ 2 ] Herein, do not forget that, the objective should be to share our findings with the readers in an easily comprehensible format. Our approach on this subject is to write all structured parts of the manuscript at the same time, and start writing the manuscript while reading the first literature. Thus newly arisen connotations, and self-brain gyms will be promptly written down. However during this process your outcomes should be revealed fully, and roughly the message of the manuscript which be delivered. Thus with this so-called ‘hunter’s approach’ the target can be achieved directly, and rapidly. Another approach is ‘collectioner’s approach. [ 3 ] In this approach, firstly, potential data, and literature studies are gathered, read, and then selected ones are used. Since this approach suits with surgical point of view, probably ‘hunter’s approach’ serves our purposes more appropriately. However, in parallel with academic development, our novice colleague ‘manuscripters’ can prefer ‘collectioner’s approach.’
On the other hand, we think that research team consisting of different age groups has some advantages. Indeed young colleagues have the enthusiasm, and energy required for the conduction of the study, while middle-aged researchers have the knowledge to manage the research, and manuscript writing. Experienced researchers make guiding contributions to the manuscript. However working together in harmony requires assignment of a chief researcher, and periodically organizing advancement meetings. Besides, talents, skills, and experiences of the researchers in different fields (ie. research methods, contact with patients, preparation of a project, fund-raising, statistical analysis etc.) will determine task sharing, and make a favourable contribution to the perfection of the manuscript. Achievement of the shared duties within a predetermined time frame will sustain the motivation of the researchers, and prevent wearing out of updated data.
According to our point of view, ‘Abstract’ section of the manuscript should be written after completion of the manuscript. The reason for this is that during writing process of the main text, the significant study outcomes might become insignificant or vice versa. However, generally, before onset of the writing process of the manuscript, its abstract might be already presented in various congresses. During writing process, this abstract might be a useful guide which prevents deviation from the main objective of the manuscript.
On the other hand references should be promptly put in place while writing the manuscript, Sorting, and placement of the references should not be left to the last moment. Indeed, it might be very difficult to remember relevant references to be placed in the ‘Discussion’ section. For the placement of references use of software programs detailed in other sections is a rational approach.
3. Which target journal should be selected?
In essence, the methodology to be followed in writing the ‘Discussion’ section is directly related to the selection of the target journal. Indeed, in compliance with the writing rules of the target journal, limitations made on the number of words after onset of the writing process, effects mostly the ‘Discussion’ section. Proper matching of the manuscript with the appropriate journal requires clear, and complete comprehension of the available data from scientific point of view. Previously, similar articles might have been published, however innovative messages, and new perspectives on the relevant subject will facilitate acceptance of the article for publication. Nowadays, articles questioning available information, rather than confirmatory ones attract attention. However during this process, classical information should not be questioned except for special circumstances. For example manuscripts which lead to the conclusions as “laparoscopic surgery is more painful than open surgery” or “laparoscopic surgery can be performed without prior training” will not be accepted or they will be returned by the editor of the target journal to the authors with the request of critical review. Besides the target journal to be selected should be ready to accept articles with similar concept. In fact editors of the journal will not reserve the limited space in their journal for articles yielding similar conclusions.
The title of the manuscript is as important as the structured sections * of the manuscript. The title can be the most striking or the newest outcome among results obtained.
Before writing down the manuscript, determination of 2–3 titles increases the motivation of the authors towards the manuscript. During writing process of the manuscript one of these can be selected based on the intensity of the discussion. However the suitability of the title to the agenda of the target journal should be investigated beforehand. For example an article bearing the title “Use of barbed sutures in laparoscopic partial nephrectomy shortens warm ischemia time” should not be sent to “Original Investigations and Seminars in Urologic Oncology” Indeed the topic of the manuscript is out of the agenda of this journal.

4. Do we have to get a pre-peer review about the written manuscript?
Before submission of the manuscript to the target journal the opinions of internal, and external referees should be taken. [ 1 ] Internal referees can be considered in 2 categories as “General internal referees” and “expert internal referees” General internal referees (ie. our colleagues from other medical disciplines) are not directly concerned with your subject matter but as mentioned above they critically review the manuscript as for simplicity, clarity, and effectiveness of its writing style. Expert internal reviewers have a profound knowledge about the subject, and they can provide guidance about the writing process of the manuscript (ie. our senior colleagues more experienced than us). External referees are our colleagues who did not contribute to data collection of our study in any way, but we can request their opinions about the subject matter of the manuscript. Since they are unrelated both to the author(s), and subject matter of the manuscript, these referees can review our manuscript more objectively. Before sending the manuscript to internal, and external referees, we should contact with them, and ask them if they have time to review our manuscript. We should also give information about our subject matter. Otherwise pre-peer review process can delay publication of the manuscript, and decrease motivation of the authors. In conclusion, whoever the preferred referee will be, these internal, and external referees should respond the following questions objectively. 1) Does the manuscript contribute to the literature?; 2) Does it persuasive? 3) Is it suitable for the publication in the selected journal? 4) Has a simple, clear, and effective language been used throughout the manuscript? In line with the opinions of the referees, the manuscript can be critically reviewed, and perfected. [ 1 ]**
Following receival of the opinions of internal, and external referees, one should concentrate priorly on indicated problems, and their solutions. Comments coming from the reviewers should be criticized, but a defensive attitude should not be assumed during this evaluation process. During this “incubation” period where the comments of the internal, and external referees are awaited, literature should be reviewed once more. Indeed during this time interval a new article which you should consider in the ‘Discussion’ section can be cited in the literature.
5. What are the common mistakes made related to the writing process of a manuscript?
Probably the most important mistakes made related to the writing process of a manuscript include lack of a clear message of the manuscript , inclusion of more than one main idea in the same text or provision of numerous unrelated results at the same time so as to reinforce the assertions of the manuscript. This approach can be termed roughly as “loss of the focus of the study” In conclusion, the author(s) should ask themselves the following question at every stage of the writing process:. “What is the objective of the study? If you always get clear-cut answers whenever you ask this question, then the study is proceeding towards the right direction. Besides application of a template which contains the intended clear-cut messages to be followed will contribute to the communication of net messages.
One of the important mistakes is refraining from critical review of the manuscript as a whole after completion of the writing process. Therefore, the authors should go over the manuscript for at least three times after finalization of the manuscript based on joint decision. The first control should concentrate on the evaluation of the appropriateness of the logic of the manuscript, and its organization, and whether desired messages have been delivered or not. Secondly, syutax, and grammar of the manuscript should be controlled. It is appropriate to review the manuscript for the third time 1 or 2 weeks after completion of its writing process. Thus, evaluation of the “cooled” manuscript will be made from a more objective perspective, and assessment process of its integrity will be facilitated.
Other erroneous issues consist of superfluousness of the manuscript with unnecessary repetitions, undue, and recurrent references to the problems adressed in the manuscript or their solution methods, overcriticizing or overpraising other studies, and use of a pompous literary language overlooking the main objective of sharing information. [ 4 ]
B) Approaches to the writing process of the ‘Discussion’ section:
1. how should the main points of ‘discussion’ section be constructed.
Generally the length of the ‘Discussion ‘ section should not exceed the sum of other sections (ıntroduction, material and methods, and results), and it should be completed within 6–7 paragraphs.. Each paragraph should not contain more than 200 words, and hence words should be counted repeteadly. The ‘Discussion’ section can be generally divided into 3 separate paragraphs as. 1) Introductory paragraph, 2) Intermediate paragraphs, 3) Concluding paragraph.
The introductory paragraph contains the main idea of performing the study in question. Without repeating ‘Introduction’ section of the manuscript, the problem to be addressed, and its updateness are analysed. The introductory paragraph starts with an undebatable sentence, and proceeds with a part addressing the following questions as 1) On what issue we have to concentrate, discuss or elaborate? 2) What solutions can be recommended to solve this problem? 3) What will be the new, different, and innovative issue? 4) How will our study contribute to the solution of this problem An introductory paragraph in this format is helpful to accomodate reader to the rest of the Discussion section. However summarizing the basic findings of the experimental studies in the first paragraph is generally recommended by the editors of the journal. [ 5 ]
In the last paragraph of the Discussion section “strong points” of the study should be mentioned using “constrained”, and “not too strongly assertive” statements. Indicating limitations of the study will reflect objectivity of the authors, and provide answers to the questions which will be directed by the reviewers of the journal. On the other hand in the last paragraph, future directions or potential clinical applications may be emphasized.
2. How should the intermediate paragraphs of the Discussion section be formulated?
The reader passes through a test of boredom while reading paragraphs of the Discussion section apart from the introductory, and the last paragraphs. Herein your findings rather than those of the other researchers are discussed. The previous studies can be an explanation or reinforcement of your findings. Each paragraph should contain opinions in favour or against the topic discussed, critical evaluations, and learning points.
Our management approach for intermediate paragraphs is “divide and conquer” tactics. Accordingly, the findings of the study are determined in order of their importance, and a paragraph is constructed for each finding ( Figure 1 ). Each paragraph begins with an “indisputable” introductory sentence about the topic to be discussed. This sentence basically can be the answer to the question “What have we found?” Then a sentence associated with the subject matter to be discussed is written. Subsequently, in the light of the current literature this finding is discussed, new ideas on this subject are revealed, and the paragraph ends with a concluding remark.

Divide and Conquer tactics
In this paragraph, main topic should be emphasized without going into much detail. Its place, and importance among other studies should be indicated. However during this procedure studies should be presented in a logical sequence (ie. from past to present, from a few to many cases), and aspects of the study contradictory to other studies should be underlined. Results without any supportive evidence or equivocal results should not be written. Besides numerical values presented in the Results section should not be repeated unless required.
Besides, asking the following questions, and searching their answers in the same paragraph will facilitate writing process of the paragraph. [ 1 ] 1) Can the discussed result be false or inadequate? 2) Why is it false? (inadequate blinding, protocol contamination, lost to follow-up, lower statistical power of the study etc.), 3) What meaning does this outcome convey?
3. What are the common mistakes made in writing the Discussion section?:
Probably the most important mistake made while writing the Discussion section is the need for mentioning all literature references. One point to remember is that we are not writing a review article, and only the results related to this paragraph should be discussed. Meanwhile, each word of the paragraphs should be counted, and placed carefully. Each word whose removal will not change the meaning should be taken out from the text.” Writing a saga with “word salads” *** is one of the reasons for prompt rejection. Indeed, if the reviewer thinks that it is difficult to correct the Discussion section, he/she use her/ his vote in the direction of rejection to save time (Uniform requirements for manuscripts: International Comittee of Medical Journal Editors [ http://www.icmje.org/urm_full.pdf ])
The other important mistake is to give too much references, and irrelevancy between the references, and the section with these cited references. [ 3 ] While referring these studies, (excl. introductory sentences linking indisputable sentences or paragraphs) original articles should be cited. Abstracts should not be referred, and review articles should not be cited unless required very much.
4. What points should be paid attention about writing rules, and grammar?
As is the case with the whole article, text of the Discussion section should be written with a simple language, as if we are talking with our colleague. [ 2 ] Each sentence should indicate a single point, and it should not exceed 25–30 words. The priorly mentioned information which linked the previous sentence should be placed at the beginning of the sentence, while the new information should be located at the end of the sentence. During construction of the sentences, avoid unnecessary words, and active voice rather than passive voice should be used.**** Since conventionally passive voice is used in the scientific manuscripts written in the Turkish language, the above statement contradicts our writing habits. However, one should not refrain from beginning the sentences with the word “we”. Indeed, editors of the journal recommend use of active voice so as to increase the intelligibility of the manuscript.
In conclusion, the major point to remember is that the manuscript should be written complying with principles of simplicity, clarity, and effectiveness. In the light of these principles, as is the case in our daily practice, all components of the manuscript (IMRAD) can be written concurrently. In the ‘Discussion’ section ‘divide and conquer’ tactics remarkably facilitates writing process of the discussion. On the other hand, relevant or irrelevant feedbacks received from our colleagues can contribute to the perfection of the manuscript. Do not forget that none of the manuscripts is perfect, and one should not refrain from writing because of language problems, and related lack of experience.
Instead of structured sections of a manuscript (IMRAD): Introduction, Material and Methods, Results, and Discussion
Instead of in the Istanbul University Faculty of Medicine posters to be submitted in congresses are time to time discussed in Wednesday meetings, and opinions of the internal referees are obtained about the weak, and strong points of the study
Instead of a writing style which uses words or sentences with a weak logical meaning that do not lead the reader to any conclusion
Instead of “white color”; “proven”; nstead of “history”; “to”. should be used instead of “white in color”, “definitely proven”, “past history”, and “in order to”, respectively ( ref. 2 )
Instead of “No instances of either postoperative death or major complications occurred during the early post-operative period” use “There were no deaths or major complications occurred during the early post-operative period.
Instead of “Measurements were performed to evaluate the levels of CEA in the serum” use “We measured serum CEA levels”
- Research Paper Guides
- Basics of Research Paper Writing
How to Write a Discussion Section: Writing Guide
- Speech Topics
- Basics of Essay Writing
- Essay Topics
- Other Essays
- Main Academic Essays
- Research Paper Topics
- Miscellaneous
- Chicago/ Turabian
- Data & Statistics
- Methodology
- Admission Writing Tips
- Admission Advice
- Other Guides
- Student Life
- Studying Tips
- Understanding Plagiarism
- Academic Writing Tips
- Basics of Dissertation & Thesis Writing
- Essay Guides
- Formatting Guides
- Basics of Research Process
- Admission Guides
- Dissertation & Thesis Guides

Table of contents
Use our free Readability checker
The discussion section of a research paper is where the author analyzes and explains the importance of the study's results. It presents the conclusions drawn from the study, compares them to previous research, and addresses any potential limitations or weaknesses. The discussion section should also suggest areas for future research.
Everything is not that complicated if you know where to find the required information. We’ll tell you everything there is to know about writing your discussion. Our easy guide covers all important bits, including research questions and your research results. Do you know how all enumerated events are connected? Well, you will after reading this guide we’ve prepared for you!
What Is in the Discussion Section of a Research Paper
The discussion section of a research paper can be viewed as something similar to the conclusion of your paper. But not literal, of course. It’s an ultimate section where you can talk about the findings of your study. Think about these questions when writing:
- Did you answer all of the promised research questions?
- Did you mention why your work matters?
- What are your findings, and why should anyone even care?
- Does your study have a literature review?
So, answer your questions, provide proof, and don’t forget about your promises from the introduction.
How to Write a Discussion Section in 5 Steps
How to write the discussion section of a research paper is something everyone googles eventually. It's just life. But why not make everything easier? In brief, this section we’re talking about must include all following parts:
- Answers for research questions
- Literature review
- Results of the work
- Limitations of one’s study
- Overall conclusion
Indeed, all those parts may confuse anyone. So by looking at our guide, you'll save yourself some hassle. P.S. All our steps are easy and explained in detail! But if you are looking for the most efficient solution, consider using professional help. Leave your “ write my research paper for me ” order at StudyCrumb and get a customized study tailored to your requirements.
Step 1. Start Strong: Discussion Section of a Research Paper
First and foremost, how to start the discussion section of a research paper? Here’s what you should definitely consider before settling down to start writing:
- All essays or papers must begin strong. All readers will not wait for any writer to get to the point. We advise summarizing the paper's main findings.
- Moreover, you should relate both discussion and literature review to what you have discovered. Mentioning that would be a plus too.
- Make sure that an introduction or start per se is clear and concise. Word count might be needed for school. But any paper should be understandable and not too diluted.
Step 2. Answer the Questions in Your Discussion Section of a Research Paper
Writing the discussion section of a research paper also involves mentioning your questions. Remember that in your introduction, you have promised your readers to answer certain questions. Well, now it’s a perfect time to finally give the awaited answer. You need to explain all possible correlations between your findings, research questions, and literature proposed. You already had hypotheses. So were they correct, or maybe you want to propose certain corrections? Section’s main goal is to avoid open ends. It’s not a story or a fairytale with an intriguing ending. If you have several questions, you must answer them. As simple as that.
Step 3. Relate Your Results in a Discussion Section
Writing a discussion section of a research paper also requires any writer to explain their results. You will undoubtedly include an impactful literature review. However, your readers should not just try and struggle with understanding what are some specific relationships behind previous studies and your results. Your results should sound something like: “This guy in their paper discovered that apples are green. Nevertheless, I have proven via experimentation and research that apples are actually red.” Please, don’t take these results directly. It’s just an initial hypothesis. But what you should definitely remember is any practical implications of your study. Why does it matter and how can anyone use it? That’s the most crucial question.
Step 4. Describe the Limitations in Your Discussion Section
Discussion section of a research paper isn’t limitless. What does that mean? Essentially, it means that you also have to discuss any limitations of your study. Maybe you had some methodological inconsistencies. Possibly, there are no particular theories or not enough information for you to be entirely confident in one’s conclusions. You might say that an available source of literature you have studied does not focus on one’s issue. That’s why one’s main limitation is theoretical. However, keep in mind that your limitations must possess a certain degree of relevancy. You can just say that you haven’t found enough books. Your information must be truthful to research.
Step 5. Conclude Your Discussion Section With Recommendations
Your last step when you write a discussion section in a paper is its conclusion, like in any other academic work. Writer’s conclusion must be as strong as their starting point of the overall work. Check out our brief list of things to know about the conclusion in research paper :
- It must present its scientific relevance and importance of your work.
- It should include different implications of your research.
- It should not, however, discuss anything new or things that you have not mentioned before.
- Leave no open questions and carefully complete the work without them.
Discussion Section of a Research Paper Example
All the best example discussion sections of a research paper will be written according to our brief guide. Don’t forget that you need to state your findings and underline the importance of your work. An undoubtedly big part of one’s discussion will definitely be answering and explaining the research questions. In other words, you’ll already have all the knowledge you have so carefully gathered. Our last step for you is to recollect and wrap up your paper. But we’re sure you’ll succeed!
How to Write a Discussion Section: Final Thoughts
Today we have covered how to write a discussion section. That was quite a brief journey, wasn’t it? Just to remind you to focus on these things:
- Importance of your study.
- Summary of the information you have gathered.
- Main findings and conclusions.
- Answers to all research questions without an open end.
- Correlation between literature review and your results.
But, wait, this guide is not the only thing we can do. Looking for how to write an abstract for a research paper for example? We have such a blog and much more on our platform.
Our academic writing service is just a click away. We are proud to say that our writers are professionals in their fields. Buy a research paper and our experts can provide prompt solutions without compromising the quality.
Discussion Section of a Research Paper: Frequently Asked Questions
1. how long should the discussion section of a research paper be.
Our discussion section of a research paper should not be longer than other sections. So try to keep it short but as informative as possible. It usually contains around 6-7 paragraphs in length. It is enough to briefly summarize all the important data and not to drag it.
2. What's the difference between the discussion and the results?
The difference between discussion and results is very simple and easy to understand. The results only report your main findings. You stated what you have found and how you have done that. In contrast, one’s discussion mentions your findings and explains how they relate to other literature, research questions, and one’s hypothesis. Therefore, it is not only a report but an efficient as well as proper explanation.
3. What's the difference between a discussion and a conclusion?
The difference between discussion and conclusion is also quite easy. Conclusion is a brief summary of all the findings and results. Still, our favorite discussion section interprets and explains your main results. It is an important but more lengthy and wordy part. Besides, it uses extra literature for references.
4. What is the purpose of the discussion section?
The primary purpose of a discussion section is to interpret and describe all your interesting findings. Therefore, you should state what you have learned, whether your hypothesis was correct and how your results can be explained using other sources. If this section is clear to readers, our congratulations as you have succeeded.

Joe Eckel is an expert on Dissertations writing. He makes sure that each student gets precious insights on composing A-grade academic writing.
You may also like


- Foundations
- Write Paper
Search form
- Experiments
- Anthropology
- Self-Esteem
- Social Anxiety

- Research Paper >
Writing a Discussion Section
Writing a discussion section is where you really begin to add your interpretations to the work.
This article is a part of the guide:
- Outline Examples
- Example of a Paper
- Write a Hypothesis
- Introduction
Browse Full Outline
- 1 Write a Research Paper
- 2 Writing a Paper
- 3.1 Write an Outline
- 3.2 Outline Examples
- 4.1 Thesis Statement
- 4.2 Write a Hypothesis
- 5.2 Abstract
- 5.3 Introduction
- 5.4 Methods
- 5.5 Results
- 5.6 Discussion
- 5.7 Conclusion
- 5.8 Bibliography
- 6.1 Table of Contents
- 6.2 Acknowledgements
- 6.3 Appendix
- 7.1 In Text Citations
- 7.2 Footnotes
- 7.3.1 Floating Blocks
- 7.4 Example of a Paper
- 7.5 Example of a Paper 2
- 7.6.1 Citations
- 7.7.1 Writing Style
- 7.7.2 Citations
- 8.1.1 Sham Peer Review
- 8.1.2 Advantages
- 8.1.3 Disadvantages
- 8.2 Publication Bias
- 8.3.1 Journal Rejection
- 9.1 Article Writing
- 9.2 Ideas for Topics
In this critical part of the research paper, you start the process of explaining any links and correlations apparent in your data.
If you left few interesting leads and open questions in the results section , the discussion is simply a matter of building upon those and expanding them.

The Difficulties of Writing a Discussion Section
In an ideal world, you could simply reject your null or alternative hypotheses according to the significance levels found by the statistics.
That is the main point of your discussion section, but the process is usually a lot more complex than that. It is rarely clear-cut, and you will need to interpret your findings.
For example, one of your graphs may show a distinct trend, but not enough to reach an acceptable significance level.
Remember that no significance is not the same as no difference, and you can begin to explain this in your discussion section.
Whilst your results may not be enough to reject the null hypothesis , they may show a trend that later researchers may wish to explore, perhaps by refining the experiment .

Self-Criticism at the Heart of Writing a Discussion Section
For this purpose, you should criticize the experiment, and be honest about whether your design was good enough. If not, suggest any modifications and improvements that could be made to the design.
Maybe the reason that you did not find a significant correlation is because your sampling was not random , or you did not use sensitive enough equipment.
The discussion section is not always about what you found, but what you did not find, and how you deal with that. Stating that the results are inconclusive is the easy way out, and you must always try to pick out something of value.
Using the Discussion Section to Expand Knowledge
You should always put your findings into the context of the previous research that you found during your literature review . Do your results agree or disagree with previous research?
Do the results of the previous research help you to interpret your own findings? If your results are very different, why? Either you have uncovered something new, or you may have made a major flaw with the design of the experiment .
Finally, after saying all of this, you can make a statement about whether the experiment has contributed to knowledge in the field, or not.
Unless you made so many errors that the results are completely unreliable, you will; certainly have learned something. Try not to be too broad in your generalizations to the wider world - it is a small experiment and is unlikely to change the world.
Once writing the discussion section is complete, you can move onto the next stage, wrapping up the paper with a focused conclusion .
- Psychology 101
- Flags and Countries
- Capitals and Countries
Martyn Shuttleworth (Mar 6, 2009). Writing a Discussion Section. Retrieved Jun 28, 2024 from Explorable.com: https://explorable.com/writing-a-discussion-section
You Are Allowed To Copy The Text
The text in this article is licensed under the Creative Commons-License Attribution 4.0 International (CC BY 4.0) .
This means you're free to copy, share and adapt any parts (or all) of the text in the article, as long as you give appropriate credit and provide a link/reference to this page.
That is it. You don't need our permission to copy the article; just include a link/reference back to this page. You can use it freely (with some kind of link), and we're also okay with people reprinting in publications like books, blogs, newsletters, course-material, papers, wikipedia and presentations (with clear attribution).
Want to stay up to date? Follow us!
Check out the official book.
Learn how to construct, style and format an Academic paper and take your skills to the next level.

(also available as ebook )
Save this course for later
Don't have time for it all now? No problem, save it as a course and come back to it later.
Footer bottom
- Privacy Policy

- Subscribe to our RSS Feed
- Like us on Facebook
- Follow us on Twitter
What’s Included: Discussion Template
This template covers all the core components required in the discussion/analysis chapter of a typical dissertation or thesis, including:
- The opening/overview section
- Overview of key findings
- Interpretation of the findings
- Concluding summary
The purpose of each section is explained in plain language, followed by an overview of the key elements that you need to cover. The template also includes practical examples to help you understand exactly what’s required, along with links to additional free resources (articles, videos, etc.) to help you along your research journey.
The cleanly formatted Google Doc can be downloaded as a fully editable MS Word Document (DOCX format), so you can use it as-is or convert it to LaTeX.
PS – if you’d like a high-level template for the entire thesis, you can we’ve got that too .
FAQ: Thesis Discussion Template
What types of dissertations/theses can this template be used for.
The discussion chapter template follows the standard format for academic research projects, which means it will be suitable for the majority of dissertations, theses and research projects (especially those within the sciences).
Keep in mind that the exact requirements for the discussion chapter/section will vary between universities and degree programs. For example, your university may require that the discussion chapter and conclusion chapter are merged into one, or that the results and discussion are covered together (this is often the case with qualitative research). So, be sure to double-check your university’s requirements before you finalise your structure.
Is this template for an undergrad, Master or PhD-level thesis?
This template can be used for a dissertation, thesis or research project at any level of study. Doctoral-level projects typically require the discussion chapter to be more extensive/comprehensive, but the structure will typically remain the same. Again, be sure to check your university’s requirements and norms in terms of document structure.
How long should the discussion chapter be?
This can vary a fair deal, depending on the level of study (undergrad, Master or Doctoral), the field of research, as well as your university’s specific requirements. Therefore, it’s best to check with your university or review past dissertations from your program to get an accurate estimate.
Can I share this template with my friends/colleagues?
Yes, you’re welcome to share this template in its original format (no editing allowed). If you want to post about it on your blog or social media, please reference this page as your source.
What format is the template (DOC, PDF, PPT, etc.)?
The dissertation discussion chapter template is provided as a Google Doc. You can download it in MS Word format or make a copy to your Google Drive. You’re also welcome to convert it to whatever format works best for you, such as LaTeX or PDF.
Do you have templates for the other chapters?
Yes, we do. We are constantly developing our collection of free resources to help students complete their dissertations and theses. You can view all of our template resources here .
Can Grad Coach help me with my discussion/analysis?
Yes, we can provide coaching-based assistance with your discussion chapter (or any other chapter). If you’re interested, get in touch to discuss our private coaching services .

Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
Pharmacological and behavioral investigation of putative self-medicative plants in Budongo chimpanzee diets
Contributed equally to this work with: Elodie Freymann, Fabien Schultz
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing
* E-mail: [email protected] (EF); [email protected] (FS)
Affiliation Primate Models for Behavioural Evolution Lab, Institute of Human Sciences, Department of Anthropology and Museum Ethnography, University of Oxford, Oxford, United Kingdom
Roles Supervision, Writing – review & editing
Affiliations Primate Models for Behavioural Evolution Lab, Institute of Human Sciences, Department of Anthropology and Museum Ethnography, University of Oxford, Oxford, United Kingdom, Gorongosa National Park, Sofala, Mozambique, Interdisciplinary Centre for Archaeology and the Evolution of Human Behaviour, University of Algarve, Faro, Portugal
Roles Funding acquisition, Supervision, Writing – review & editing
Affiliations Ethnopharmacology & Zoopharmacognosy Research Group, Department of Agriculture and Food Sciences, Neubrandenburg University of Applied Sciences, Neubrandenburg, Germany, ZELT–Center for Nutrition and Food Technology gGmbH
Roles Formal analysis, Writing – original draft, Writing – review & editing
Affiliation Ethnopharmacology & Zoopharmacognosy Research Group, Department of Agriculture and Food Sciences, Neubrandenburg University of Applied Sciences, Neubrandenburg, Germany
Roles Resources, Supervision, Writing – review & editing
Affiliations Wild Minds Lab, School of Psychology and Neuroscience, University of St Andrews, St Andrews, United Kingdom, Budongo Conservation Field Station, Masindi, Uganda
Affiliation Wildlife Research Center, Inuyama Campus, Kyoto University, Inuyama, Japan
Roles Investigation
Affiliation Budongo Conservation Field Station, Masindi, Uganda
Roles Formal analysis
Affiliations Budongo Conservation Field Station, Masindi, Uganda, Czech University of Life Sciences Prague, Prague, Czech Republic
Roles Resources, Writing – review & editing
Affiliations Budongo Conservation Field Station, Masindi, Uganda, Department of Comparative Cognition, Institute of Biology, University of Neuchâtel, Neuchâtel, Switzerland
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing
Affiliations Ethnopharmacology & Zoopharmacognosy Research Group, Department of Agriculture and Food Sciences, Neubrandenburg University of Applied Sciences, Neubrandenburg, Germany, Pharmacognosy and Phytotherapy, School of Pharmacy, University College of London, London, United Kingdom
- Elodie Freymann,
- Susana Carvalho,
- Leif A. Garbe,
- Dinda Dwi Ghazhelia,
- Catherine Hobaiter,
- Michael A. Huffman,
- Geresomu Muhumuza,
- Lena Schulz,
- Daniel Sempebwa,

- Published: June 20, 2024
- https://doi.org/10.1371/journal.pone.0305219
- Reader Comments
Wild chimpanzees consume a variety of plants to meet their dietary needs and maintain wellbeing. While some plants have obvious value, others are nutritionally poor and/or contain bioactive toxins which make ingestion costly. In some cases, these nutrient-poor resources are speculated to be medicinal, thought to help individuals combat illness. In this study, we observed two habituated chimpanzee communities living in the Budongo Forest, Uganda, and collected 17 botanical samples associated with putative self-medication behaviors (e.g., bark feeding, dead wood eating, and pith-stripping) or events (e.g., when consumer had elevated parasite load, abnormal urinalysis, or injury). In total, we selected plant parts from 13 species (nine trees and four herbaceous plants). Three extracts of different polarities were produced from each sample using n -hexane, ethyl acetate, and methanol/water (9/1, v/v ) and introduced to antibacterial and anti-inflammatory in vitro models. Extracts were evaluated for growth inhibition against a panel of multidrug-resistant clinical isolates of bacteria, including ESKAPE strains and cyclooxygenase-2 (COX-2) inhibition activity. Pharmacological results suggest that Budongo chimpanzees consume several species with potent medicinal properties. In the antibacterial library screen, 45 out of 53 extracts (88%) exhibited ≥40% inhibition at a concentration of 256 μg/mL. Of these active extracts, 41 (91%) showed activity at ≤256μg/mL in subsequent dose-response antibacterial experiments. The strongest antibacterial activity was achieved by the n- hexane extract of Alstonia boonei dead wood against Staphylococcus aureus (IC50: 16 μg/mL; MIC: 32 μg/mL) and Enterococcus faecium (IC50: 16 μg/mL; MIC: >256 μg/mL) and by the methanol-water extract of Khaya anthotheca bark and resin against E . faecium (IC50: 16 μg/mL; MIC: 32 μg/mL) and pathogenic Escherichia coli (IC50: 16 μg/mL; MIC: 256 μg/mL). We observed ingestion of both these species by highly parasitized individuals. K . anthotheca bark and resin were also targeted by individuals with indicators of infection and injuries. All plant species negatively affected growth of E . coli . In the anti-inflammatory COX-2 inhibition library screen, 17 out of 51 tested extracts (33%) showed ≥50% COX-2 inhibition at a concentration of 5 μg/mL. Several extracts also exhibited anti-inflammatory effects in COX-2 dose-response experiments. The K . anthotheca bark and resin methanol-water extract showed the most potent effects (IC50: 0.55 μg/mL), followed by the fern Christella parasitica methanol-water extract (IC50: 0.81 μg/mL). This fern species was consumed by an injured individual, a feeding behavior documented only once before in this population. These results, integrated with associated observations from eight months of behavioral data, provide further evidence for the presence of self-medicative resources in wild chimpanzee diets. This study addresses the challenge of distinguishing preventative medicinal food consumption from therapeutic self-medication by integrating pharmacological, observational, and health monitoring data—an essential interdisciplinary approach for advancing the field of zoopharmacognosy.
Citation: Freymann E, Carvalho S, Garbe LA, Dwi Ghazhelia D, Hobaiter C, Huffman MA, et al. (2024) Pharmacological and behavioral investigation of putative self-medicative plants in Budongo chimpanzee diets. PLoS ONE 19(6): e0305219. https://doi.org/10.1371/journal.pone.0305219
Editor: Armel Jackson Seukep, University of Buea, CAMEROON
Received: January 9, 2024; Accepted: May 25, 2024; Published: June 20, 2024
Copyright: © 2024 Freymann et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All relevant data are within the manuscript and its Supporting Information files.
Funding: Funding for this project was granted by the the Clarendon Fund at the University of Oxford (to EF), the British Institute of Eastern Africa (to EF), Keble College at the University of Oxford (to EF), Boise Trust Fund (to EF), German Federal Ministry of Education and Research (13FH026IX5, PI: L-AG and Co-I: FS) (to LAG, FS) and Neubrandenburg University of Applied Sciences (grant # 13310510) (to LAG, FS).
Competing interests: The authors have declared that no competing interests exist.
Introduction
‘Medicinal foods’ refer to resources in the diet that have potential curative value due to the presence of plant secondary metabolites (PSMs) [ 1 , 2 ]. PSMs are compounds that usually occur only in special, differentiated cells [ 3 ] and which help plants defend against predators, pathogens, and competitors [ 4 – 7 ]. PSMs can have a range of functions, including the inhibition of microbial, fungal, and competitor growth [ 8 ]. While some PSMs can be toxic at high doses, these compounds can also promote the health of human and non-human consumers [ 8 – 10 ]. Research suggests 15–25% of primate and other mammalian diets consist of medicinal foods [ 9 , 11 ]. These resources likely play a critical role in animal health-maintenance by passively preventing or reducing the impact of parasitic infections or other pathogens [ 9 – 14 ].
While most animals likely consume foods with medicinal properties as part of their normal diets, fewer species have been shown to engage in therapeutic self-medication. Huffman [ 15 ] defines this type of self-medicative behavior as the active extraction and ingestion, by an ill individual, of medicinal resources with little nutritional value. Instead of an individual passively benefiting from a plant’s medicinal properties through normal feeding, this form of self-medication requires basic awareness of the resource’s healing properties. One of the best-studied animals to engage in this form of self-medication is our closest living relative: the chimpanzee.
Wild chimpanzees ( Pan troglodytes ), across at least sixteen field sites [ 15 ] have demonstrated therapeutic self-medication using two well-established self-medicative behaviors: leaf swallowing [ 16 , 17 ] and bitter-pith chewing [ 18 ]. Leaf swallowing, first reported by Wrangham [ 19 , 20 ] and described by Wrangham & Nishida [ 21 ], involves the careful selection and ingestion of whole, hispid leaves. This behavior was later demonstrated to expel internal parasites (i.e. Oesophagostomum sp. and Bertiella studeri ) from the gut [ 16 , 17 , 22 , 23 ]. The functional mechanism responsible for this anthelminthic effect is considered to be primarily “mechanical” [ 9 ] as, rather than a chemical compound, the leaf’s indigestibility, brought about by the trichomes on its surface—stimulates gut motility in the swallower [ 17 , 23 , 24 ].
The second established behavior is bitter-pith chewing, which involves the stripping of outer bark and leaves from the soft new stem growth of the shrub, Vernonia amygdalina , exposing the inner pith. Individuals chew the pith and ingest only the bitter juices while spitting out the fibers [ 18 , 25 ]. Bitter-pith chewing is considered ‘phytochemical’ self-medication [ 9 ], as its anthelminthic effect appears to be the result of bioactive PSMs [ 26 – 29 ]. This behavior’s medicinal effect was associated with a significant drop in the infection intensity of Oesophagostomum stephanostomum nematodes [ 25 ], suggesting that the bitter compounds directly affect the adult worms. This hypothesis was supported by in vivo studies conducted by Jisaka et al. [ 30 ], demonstrating that extracts from the pith permanently paralyzed adult Schistosome parasites. V . amygdalina is also used to aid gastrointestinal discomfort and other signs of parasitosis in humans and livestock, symptoms also displayed by chimpanzees ingesting the plant’s bitter pith [ 9 , 18 , 25 , 31 ]. The bitter piths of other plant species are reported to be chewed by chimpanzees across field sites but detailed studies on their medicinal properties have yet to be conducted [ 9 ].
Beyond these two established behaviors, not much is known about the phytochemical self-medicative repertoires of wild chimpanzees, although some behaviors associated with the ingestion of specific plant parts or processing techniques have been recommended for further investigation [ 9 , 15 , 32 ]. One of these behaviors is bark feeding, which involves the ingestion of living stem bark and/or cambium [ 33 ], and which has been observed in at least eleven established field sites [ 33 – 43 ]. Bark feeding has been suggested as a medicinal behavior in chimpanzees and other primates, used to aid in the chemical control of intestinal nematode infection and to relieve gastrointestinal upset [ 9 ]. Bark is characteristically highly fibrous, heavily lignified, sometimes toxic, relatively indigestible, and nutrient-poor [ 44 ]. However, the contribution of bark in chimpanzee diets and toward general health is still poorly understood [though see: 45 ]. In this study, the bark of eight species ingested by Budongo chimpanzees ( Scutia myrtina , Cynometra alexandri , Alstonia boonei , Ficus exasperata , Ficus variifolia , Syzygium guineense , Desplatsia dewevrei , Khaya anthotheca) was screened for antibiotic and anti-inflammatory properties, to better understand the function of bark feeding behaviors and the role this behavior may play in the health maintenance of chimpanzees. For the species K . anthotheca , we tested a mixture of bark and congealed resin, which Budongo chimpanzees were observed to particularly target throughout the study period.
Another putative self-medicative behavior is dead wood eating [ 9 , 35 ], which involves the consumption of decomposing cambium from dead trees. To date, the majority of studies examining this behavior in apes have focused on exploring potential mineral and nutritional benefits, rather than investigating pharmacological properties [ 46 – 49 ]. Many of these studies suggest that dead wood is exploited by chimpanzees as a source of sodium in environments where this mineral is otherwise scarce [ 48 , 49 ]. Our study evaluates the pharmacology of two species of dead wood ( A . boonei and Cleistopholis patens) consumed by the Sonso community of chimpanzees to determine whether this behavior may have multiple functions or health benefits.
The ingestion of pith material from other species has also been suggested as putatively self-medicative [ 34 , 50 , 51 ]. However, unlike V . amygdalina bitter-pith, some of these plant piths appear bland or tasteless. While Wrangham et al. have previously suggested that pith is likely a high-fiber fallback food [ 52 ], De la Fuente et al. review several pith species targeted by chimpanzees with proposed medicinal properties [ 32 ]. In our study, two species of non-bitter piths ( Marantachloa leucantha and Acanthus polystachyus) , were collected for pharmacological assessment. M . leucantha was observed on several occasions being stripped, masticated, and spat out after the juice was extracted from the pith, whereas A . polystachyus was observed being stripped, masticated, and swallowed. Both of these species are also ingested by chimpanzees in Kibale National Park, Uganda [ 52 ].
Establishing phytochemical self-medicative behaviors in wild animals is difficult and time consuming, as the burden of proof is high, self-medicative events can be rare relative to other behaviors, and methods often require multidisciplinary expertise and collaboration [ 9 ]. Past studies have utilized ethnopharmacological methods to determine specific medicinal properties of foods consumed by primates [ 11 ], greatly advancing our understanding of the relationship between primate diets and health. However, a key challenge for establishing novel self-medicative behaviors is differentiating between medicinal food consumption and therapeutic self-medication. While pharmacological data interpreted on its own is crucial for establishing the presence of medicinal resources in chimpanzee diets, the integration of observational and health monitoring data is needed to parse therapeutic self-medicative behaviors from normal feeding behaviors with inadvertent health benefits. Furthermore, the importance of collecting in situ samples from the locations where putative self-medicative behaviors are observed is paramount, as ecological, climatic, and anthropogenic variables can cause variation in the bioactivity of plants across habitats [ 53 ].
In total, we investigated the bioactivity of 51 plant extracts produced from 17 part-specific samples (across 13 species), collected in the Budongo Forest. Each extract was tested for inhibition of bacterial growth as well as anti-inflammatory COX-2 inhibition activity. Due to limitations in scope, funding, and the unavailability of anthelminthic assays for wild animal parasites, none were not conducted in this study, restricting specific identification of parasiticidal behaviors. Assay results are reported and contextualized in this study with direct behavioral evidence and health monitoring data.
Materials and method
Study site and subjects.
Behavioral data, health monitoring metrics, and botanical samples were collected from the Budongo Central Forest Reserve in Uganda (1°35′– 1°55′ N, 31°18′–31°42′ E). An overview of methodological workflow can be found in S2 Fig . The Budongo Conservation Field Station (BCFS) site, founded in 1990, is composed of continuous, semi-deciduous forest and contains two habituated Eastern chimpanzee ( Pan troglodytes schweinfurthii ) communities [ 54 ]. The Sonso community has been studied continuously since 1992, and the ages, social relationships, demographics, and diet of its members are well documented [ 55 , 56 ]. The Sonso population was ~68 individuals at the time of data collection, and the home range covered an area of ~5.33 km 2 [ 57 ]. Waibira, a larger group of at least 105 individuals, was more recently habituated, with consistent data collection beginning in 2011. The Waibira maximum home range area was ~10.28 km 2 [ 57 ].
Behavioral data collection
All samples were collected in the Budongo Forest within the Sonso home range, based on behavioral observations from the study period and supporting evidence from the site’s long-term data of their use. Behavioral and health data were collected from two neighboring chimpanzee communities, each for one four-month field season (Sonso: June-October 2021, Waibira: June-October 2022). Data collected between June-September 2021 informed subsequent plant sample collection for pharmacological analysis, which occurred in early September 2021. Behavioral data collected after sample collection provided additional behavioral context for ingestion of these species. Behavioral data were collected between 07:00 and 16:30 in Sonso and between 06:30 and 17:00 in Waibira using day-long focal animal follows sensu Altman et al. [ 58 ]. This data was recorded using Animal Observer (AO) on iPad and ad libitum feeding events were recorded for any unusual feeding behaviors, including but not limited to bark ingestion, dead wood eating, pith stripping, and geophagy. All feeding events were filmed on a Sony Handycam CX250. We prioritized focal follows on individuals with wounds, high or diverse parasite loads identified through on-going monitoring, or known ailments. However, consecutive day follows of priority individuals were not always possible—or were avoided when they might contribute to increased stress in particularly vulnerable individuals. Throughout the study, using this protocol, 27 Sonso individuals (♂:11; ♀:16) and 24 Waibira individuals (♂:14; ♀:10) were observed. Authors collecting behavioral data were blind to pharmacological results during both study periods.
Health monitoring
Individual health data were recorded in both communities, including opportunistic macroscopic and microscopic fecal analysis and urinalysis testing. While anthelminthic assays were not run in this study, parasite load was opportunistically assessed to provide additional health context for each observation. As the presence of certain helminths may impair a host’s immunological response to bacterial, viral, and protozoal pathogens [ 59 ], parasite load can provide a proxy measurement for overall health. Similarly, a reduced immune system and increased stress caused by co-infections could render a host more susceptible to virulent endoparasites [ 60 , 61 ]. When helminths and/or proglottids were found in samples, they were collected and preserved in ethanol for later identification. To quantify parasite loads, fecal samples were analyzed using the McMaster Method [ 9 , 25 , 62 ]. Urinalysis samples were taken opportunistically using multi-reagent Urine Dipstick Test 9-RC for Urotron RL9 to assess the health and physiological status of group members following methods established by Kaur & Huffman [ 63 ]. Urinalysis metrics considered in this study included: leukocytes (LEU) associated with pyuria caused by UTI, balanitis, urethritis, tuberculosis, bladder tumors, viral infections, nephrolithiasis, foreign bodies, exercise, glomerulonephritis, and corticosteroid and cyclophosphamide use; blood (BLO) associated with peroxidase activity of erythrocytes, and UTIs; and ketones (KET) associated with pregnancy, carbohydrate-free diets, starvation, and diabetes [ 64 ]. Test results were interpreted in situ using a colorimetric scale. We considered a result ‘abnormal’ if the colorimetric scale indicated a positive result when the expected result was negative or if the result was outside the specified test parameters according to the manufacturer.
Plant sample selection for bioactivity testing
Plants were selected for pharmacological testing after three months of data collection in the Sonso community. We selected 10 samples (from 9 species) based on direct observations during this period. These observations included individuals targeting plant parts associated with putative self-medicative behaviors (i.e., bark feeding, dead wood eating, pith-stripping) or sick/wounded individuals seeking out unusually consumed resources. We then selected an additional five species, the ingestion of which had not been directly observed, for testing based on their historical inclusion in Sonso chimpanzees’ bark feeding repertoire. GM, who has worked at the field station for over thirty-years, has previously observed bark feeding on each of these selected species. These historic observations enabled collection of bark samples from specific trees known to have been previously stripped. In two cases, leaf samples were collected from tree species that were also selected for bark samples ( S . guineense and F . exasperata) . While neither Sonso nor Waibira chimpanzees have been observed ingesting the leaves of S . guineense , a sample was collected to enable comparison of bioactivity across plant parts. F . exasperata leaves are consumed in both communities; however, we found no behavioral evidence for use in unusual contexts. In some cases, direct observation of an event involving one of the collected species occurred after botanical collection was complete. These post hoc behavioral observations are reported in this paper, although they did not impact sample selection.
Collection of sample material
Plants were collected from the Sonso community home range following best practice procedures [ 65 ], using sustainable harvesting methods [ 66 ]. See S1 File for more information. Voucher accession numbers are reported in Table 3 . Digital images of voucher specimens can be found in S3 Fig . The currently recognized scientific names of each species were confirmed on https://mpns.science.kew.org/ . Plant family assignments were done in accordance with The Angiosperm Phylogeny Group IV guidance [ 67 ].
Ethnobotanical literature review
We conducted a post-hoc ethnomedicinal review of all species collected for this study using Google Scholar, PROTA, and Kokwaro’s ethnomedicinal pharmacopeia [ 68 ]. To search databases, we used scientific names and synonyms for each plant as keywords [ 65 ].
Plant processing and extractions
At Neubrandenburg University of Applied Sciences, samples were ground using a food processor. Extractions were produced using two solvents and a solvent mixture ( n -hexane, ethyl acetate, and methanol/water ( v/v 9/1)), allowing for the selective isolation of components with varying solubilities and polarities. Methanol-water, the solvent with the highest polarity, generally extracts primary plant metabolites (e.g., polar compounds such as proteins, amino acids, and carbohydrates). Nonpolar solvents like n- hexane extract nonpolar compounds like lipids, making n-hexane a preferred solvent for oil or wax extraction. Extractions with each solvent were achieved through double maceration of new material (non-successively). Extraction suspensions were placed on a shaker at 80 rpm at room temperature for minimum 72h, followed by vacuum filtration. Processes were repeated with the leached material. Filtrates were then combined and dried using a vacuum evaporator, labeled, and stored at -20°C until needed for assays.
Sample solution preparation
To create sample solutions, each crude extract was dissolved in DMSO (Carl Roth) at a concentration of 10 mg/mL. To ensure a homogenous solution, samples were mixed with a vortex mixer and, if necessary, treated with sonication at room temperature or up to 55°C for samples with low solubility. Each extract solution was then tested for inhibition of bacterial growth as well as anti-inflammatory COX-2 inhibition activity. Solutions were stored at -20°C when not in use.
Antibacterial susceptibility tests
A. bacterial strains..
For antibacterial assays, eleven multidrug-resistant clinical isolate strains from nine species were used. This process increased the study’s applicability for early-stage drug discovery, specifically relevant to the threat of antimicrobial resistance (AMR). Seven of these strains (from six species) are classified as ESKAPE pathogens, including Enterococcus faecium (DSM 13590), Staphylococcus aureus (DSM 1104; DSM 18827), Klebsiella pneumoniae (DSM 16609), Acinetobacter baumannii (DSM 102929), Pseudomonas aeruginosa (DSM 1117), and Enterobacter cloacae (DSM 30054), meaning they are highly virulent and resistant to antibiotics [ 69 ]. A strain of the foodborne pathogen Escherichia coli (DSM 498) with AMR as well as a non-resistant E . coli strain (DSM 1576) were also included in the study. Although not an ESKAPE pathogen, E . coli is widely known for causing bacterial diarrhea and AMR strains are a major cause of urinary tract infections [ 70 , 71 ]. Strains of Stenotrophomonas maltophilia (DSM 50170) and Salmonella enterica subsp. enterica (DSM 11320) were also tested. More information on specific clinical isolates/strains, their individual resistance profiles, and antibiotics used can be found in the S5 & S6 Tables in S2 File . Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution testing (M100-S23) were followed [ 72 ].
b. Growth inhibition screening and dose-response study.
The broth dilution in vitro methods for bacterial susceptibility assessment have previously been described by Schultz et al. [ 69 ]. The standardized bacterial working cultures were pipetted into sterile 96-well microtiter plates (Greiner Bio-One International, CELLSTAR 655185). Extracts and antibiotic (64–1 μg/mL), vehicle and sterility controls, were then added into respective wells. Initial optical density measurement (600 nm) was performed, accounting for absorbance of extracts. Plates were incubated at 37°C for 18 h, except for A . baumannii which was incubated for 22h in accordance with strain characteristics ( S5 Table in S2 File ) . After incubation, a final optical density reading (600 nm) was conducted. Percent inhibition values were calculated and the IC 50 and MIC values were determined [ 69 , 73 ]. The IC 50 value is defined as the lowest concentration at which an extract showed ≥ 50% inhibition, and the MIC is the lowest concentration at which an extract displayed ≥ 90% inhibition. A total of 51 samples underwent single-dose pre-screening for growth inhibition (in triplicate) at the concentration of 256 μg/mL on eleven pathogens. Samples showing ≥40% growth inhibition were further tested in a dose-response study with two-fold serial dilution at descending concentrations from 256 to 4 μg/mL. The dose-response experiments were done as biological replicates on separate days in triplicate (technical replicates) to validate reproducibility. Positive controls (antibiotics) and negative controls (vehicle control and sterile media control) were always included. Further details on bacteria standardization can be found in S1 File . Information on plate setup for bacterial library screens and dose-response assays can be found in S4 Fig .
COX-2 inhibition assay
Anti-inflammatory assays were assessed using an in vitro COX inhibitor screening assay kit (Cayman Item No: 701080), with modifications previously described in Schultz et al. [ 74 ]. All extracts were first screened in duplicate for inhibition against human recombinant COX-2 at an initial concentration of 50 μg/mL. For extracts exhibiting at least 50% inhibition, the concentration was then lowered to 10 μg/mL, 5 μg/mL, and 2.5 μg/mL. The most active extracts were taken to dose-response experiments for determination of IC 50 values ( Table 5 ). The assay was done in two steps: 1) the COX reaction step in which the prostaglandin H 2 (PG) was produced (which was further reduced to the more stable prostaglandin F 2α by addition of stannous chloride), and 2) an acetyl choline esterase competitive ELISA step to quantify the produced prostaglandin and calculate a potential enzyme inhibition caused by the extracts. The pure compound and selective COX-2 inhibitor DuP-769 was included as a positive control. DMSO was included as the vehicle control for determining 100% enzyme activity. Information on ELISA plate setup for anti-inflammation assays can be found in S5 Fig .
Ethics statements
Behavioral data used in this study were collected with the approval of the Uganda Wildlife Authority (permit #: COD/96/05) and the Uganda National Council for Science and Technology (permit #: NS257ES). Exportation of samples for pharmacological testing were conducted under UNCST permit #: NS104ES. Behavioral data collection adhered to International Primatological Society’s Code of Best Practice for Field Primatology [ 75 ]. No exported samples were listed under CITES. Plant samples were exported in collaboration with Makerere University (permit #: UQIS00005033/93/PC), issued by the Ugandan government, and transported to Neubrandenburg University of Applied Sciences in accordance with the Nagoya Protocol. A CUREC was approved by the University of Oxford (Ref No.: SAME_C1A_22_080). The authors report no conflict of interest.
Behavioral observations
Several unusual feeding events and putative self-medicative behaviors were recorded over 116 total field days. Table 1 reports all species collected for pharmacological testing and provides behavioral justifications for collection. Images from some of these events can be found in S1 Fig .
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pone.0305219.t001
Individuals with injuries were directly observed ingesting K . anthotheca bark and resin, W . elongata young leaves, C . alexandri bark, and C . parasitica ferns. Individuals exhibiting respiratory symptoms were observed ingesting C . alexandri bark and K . anthotheca bark and resin. Individuals with abnormal urinalysis results (e.g., positive for leukocytes, elevated ketones, and presence of blood) were observed feeding on C . patens dead wood, K . anthotheca bark and resin, and M . leucantha pith. Individuals with recent cases of diarrhea were observed consuming A . boonei and C . patens dead wood, K . anthotheca bark and resin, and W . elongata leaves. Parasitological analyses further suggest individuals with varying degrees of endoparasite infections consumed S . myrtina and C . alexanderi bark, A . boonei and C . patens dead wood, K . anthotheca bark and resin, W . elongata leaves, as well as A . polystachyus and M . leucantha pith. On a day when two individuals were observed leaf swallowing, a scientifically established self-medicative behavior, one was observed consuming K . anthotheca bark and resin, while the other was observed stripping A . polystachyus pith prior to the event. Ingestion of F . variifolia , D . dewevrei , and S . guineense bark were never directly observed during the study period. Examples of bark feeding, dead wood eating, and pith-stripping marks are shown in Fig 1 .
[ a ]: Evidence of F. exasperata bark feeding [ b ] Evidence of C. patens dead wood eating [ c ] Evidence M. leucantha pith-stripping and wadging.
https://doi.org/10.1371/journal.pone.0305219.g001
Ethnobotanical review
Based on our analysis of ethnomedicinal literature spanning various African regions from 1976 to 2022, 11 out of the 13 species tested also had documented ethnomedicinal uses ( Table 2 ).
https://doi.org/10.1371/journal.pone.0305219.t002
Production of extracts and sample information
Taxonomic information and extraction details for the 13 plant species studied, including the plant family, local name (when available), plant part used, solvent for extraction, yield of extraction, extract identification numbers (extract IDs), herbarium accession numbers, and collection location are summarized in Table 3 . Overall, the highest extraction yields were obtained with methanol-water (9/1) as a solvent. The yields from methanol-water extractions for C . parasitica , F . exasperata leaves, and S . guineense stem bark were higher than the other extractions from these samples. The plant samples which had higher yield values with n -hexane, such as the leaves of W . elongata and bark extract of A . boonei , likely have a higher content of lipids (i.e., fatty molecules).
https://doi.org/10.1371/journal.pone.0305219.t003
Library screening against multidrug-resistant human and food bacterial pathogens
Initial screening of extracts involved checking for growth inhibition against each bacterium at a concentration of 256 μg/mL. In total, 45 of the 51 plant extracts (88%) showed activity ≥40% inhibition against at least one of the 11 strains and were thus considered active and brought to dose-response experiments to determine their IC 50 value and MIC. Results from the library screening are reported in S1 Table in S2 File . As all tested plant species in the library screen had at least one extract that was active ( in vitro ) against at least one bacterial strain, no entire species was eliminated for further experimentation. However, as no extracts (at any concentration) inhibited the growth of K . pneumoniae , no further tests were conducted on this bacterium. The extract active against the most bacterial strains (n = 11) was the methanol-water extract of S . guineense stem bark (mwE098a, active against eight strains), followed by the methanol-water S . guineense leaves (mwE098b), the ethyl acetate P . patens dead wood, and the n -hexane A . boonei dead wood (hE092b) extracts, which were each active against seven, seven, and six strains, respectively. The only extract that demonstrated significant inhibition against P . aeruginosa at the highest test concentration was the methanol-water extract from S . guineense bark (mwE098a). This was also the only extract to display significant inhibition at 256 μg/mL against E . cloacae . Of all bacteria in this study, the two strains of E . coli (DSM 498 and DSM 15076) were the most susceptible, with at least one extract from all plant species inhibiting their growth. The E . coli strain with nine known antibiotic resistances (DSM 15076) surprisingly showed growth inhibition in 80% of tested extracts.
Dose-response antibacterial experiments
In dose-response assays, 41 out of the 45 tested extracts (91%) showed activity at ≤256μg/mL, though not all extracts reached MIC values (see Table 4 ). The results, along with standard deviations, are reported in S2 Table in S2 File , while S3 Table in S2 File provides a summary of the number of strains each extract was active against. The strongest in vitro growth inhibition was reported for the methanol-water extract of K . anthotheca bark and resin (mwE088) against Gram-positive E . faecium and the n- hexane extract of A . boonei dead wood (hE092b) against Gram-positive S . aureus (DSM 1104). Both extracts had low IC 50 values of 16 μg/mL (showing strong inhibition), with MIC values of 32 μg/mL against respective strains. E . faecium showed the most general susceptibility to K . anthotheca , with all extracts of this species achieving MIC values (mwE088: 32 μg/mL, eE088: 64 μg/mL, hE088: 128 μg/mL). The ethyl acetate extract of A . boonei dead wood (eE092b) also strongly inhibited the growth of E . faecium (IC 50 : 16 μg/mL; MIC: 64 μg/mL), as did the n- hexane extract of A . boonei dead wood, producing an IC 50 value of 16 μg/mL but failing to reach a MIC value. S . aureus (DSM 1104) was also highly susceptible to the ethyl acetate extracts of A . boonei dead wood (IC 50 : 32 μg/mL; MIC: 128 μg/mL).
https://doi.org/10.1371/journal.pone.0305219.t004
Only one extract, the methanol-water extract of S . guineense bark (mwE098a), was active against the gram-negative P . aeruginosa . This extract exhibited moderate growth inhibition (IC 50 : 64 μg/mL) with no MIC value reached. Despite E . coli (DSM 498) being highly susceptible on the library screen, only two extracts, the methanol-water extract of A . boonei dead wood (mwE092b; IC 50 : 256 μg/mL) and the methanol-water extract of S . guineense leaves (mwE098b; IC 50 : 128 μg/mL), reached IC 50 values at the concentration range tested, with no MICs reached. Interestingly, the strain of E . coli with nine known resistances (DSM 1576) was more susceptible, with 89% (N = 40) of extracts achieving IC 50 values ≤ 256 μg/mL. The most active extract against this strain was the methanol-water extract of K . anthotheca (mwE088; IC 50 : 16 μg/mL; MIC: 256 μg/mL). S . guineense exhibited the highest overall inhibition of S . maltophilia , with all extracts except hE098a displaying IC 50 values of ≤ 256 μg/mL against the bacterium. At the concentration range tested, no extracts yielded MIC values for S . aureus (DSM 18827), A . baumannii , E . cloacae , P . aeruginosa or E . coli (DSM 498).
Anti-inflammatory COX-2 inhibition library screen
Results from the in vitro COX-2 inhibition library screen at descending concentrations are reported in S4 Table in S2 File . At the initial concentration of 50 μg/mL, 43 out of 51 extracts (84%) exhibited an enzyme inhibition of at least 50%, displaying anti-inflammatory activity. This included at least one extract of every plant species. In the next stage of screening, at 10 μg/mL, 18 samples were eliminated. During the final step, at 5 μg/mL, five more were eliminated. The remaining 17 extracts from 10 plant species which displayed inhibition ≥50% at 5 μg/mL, were then introduced to dose-response experiments. The ethyl acetate S . myrtina bark extract (eE089b) was taken to the COX-2 dose-response despite not showing inhibition past 50 μg/mL, as it almost reached the selection limit during analysis and had a relatively high standard deviation. No extracts from W . elongata , C . patens or D . dewevrei showed COX-2 inhibition at 5 μg/mL and thus were excluded from further testing.
COX-2 inhibition dose-response experiments
The most active COX-2 inhibitors were extracts from K . anthotheca (mwE088; hE088; eE088), C . parasitica (mwE087; hE087), F . exasperata (hE093a; eE093a), S . myrtina (hE089a; eE089b), F . variifolia (eE097; hE097), A . polystachyus (hE099; eE099), M . leucantha (hE094), S . guineense (hE098a), A . boonei (hE092b), and C . alexandri (hE096). Results are reported in Table 5 . The strongest COX-2 inhibitor was the K . anthotheca methanol-water bark and resin extract (mwE088) (IC 50 of 0.55 μg/mL), followed by the C . parasitica methanol-water fern extract (mwE087) (IC 50 of 0.81 μg/mL). In contrast, all extracts of the species W . elongata , C . patens , and D . dewevrei failed to show ≥50% inhibition, mostly at the second screening concentration (10 μg/mL). W . elongata extracts notably showed low activity in both antibacterial and COX-2 inhibition assays.
https://doi.org/10.1371/journal.pone.0305219.t005
Plant species with strong pharmacological activity
This study provides the first pharmacological and behavioral evidence of its kind, based on in situ sampling, for the medicinal benefits of bark feeding, dead wood eating, and non-bitter pith stripping behaviors in Budongo chimpanzees. In the following sub-sections, we describe and discuss specific results from five of the tested plant species in further detail. For scope, we selected the two species with the strongest antibacterial properties ( K . anthotheca and A . boonei ) to profile, both of which were the only species to reach 40% inhibition at 16 μg/mL. We also selected C . parasitica to discuss as this species, along with K . anthotheca , exhibited the strongest anti-inflammatory properties. We then discuss results from our S . guineense samples, as this species was effective against the most bacterial strains in our antibacterial assays. Lastly, we selected S . myrtina , as we have behavioral evidence and health data that anecdotally support the use of this species for therapeutic self-medication by Budongo chimpanzees.
Alstonia boonei . Numerous in vitro and in vivo studies, reviewed by Adotey [ 76 ], have reported pharmacological activity in A . boonei bark. However, none of these studies investigated dead wood samples of A . boonei . Consistent with these findings, we found high levels of antibacterial and anti-inflammatory activity in the extracts of this species. Interestingly, extracts from A . boonei dead wood generally exhibited higher activity than living bark. This difference could be due either to a change in active ingredient composition, or possible fungal growth following the tree’s death. While the A . boonei dead wood n -hexane extract (hE092b) exhibited strong growth inhibition against S . aureus (DSM 1104; DSM 18827) and E . faecium at low concentrations in the dose-response assays, the n -hexane bark extract (hE092a) showed no activity <256 μg/mL. Similarly, the ethyl acetate extract of dead wood (eE092b) also strongly inhibited S . aureus (DSM 1104) (IC 50 : 16 μg/mL; MIC: 128 μg/mL) and E . faecium (IC 50 : 16 μg/mL; MIC: 64 μg/mL), while the ethyl acetate bark extract of this species did not even exhibit enough inhibition in the antibacterial library screen to be taken to dose-response assays. However, the methanol-water extract of A . boonei bark (mwE092a) did show activity against E . coli (DSM 498) (IC 50 : 128 μg/mL), as did the methanol-water dead wood extract (mwE092a) (IC 50 : 128 μg/mL), with no MIC values reached in either case. Overall, extracts from A . boonei displayed more potent activity in Gram-positive bacteria, although this effect is more apparent in dead wood than stem bark. In the COX-2 inhibition assays, the n -hexane extract of A . boonei dead wood also showed strong anti-inflammatory inhibition, while the n -hexane extract of the bark only exhibited weak inhibition (at the highest test concentration of 50 μg/mL).
A . boonei is a known medicinal plant across East Africa, commonly used for a variety of reproductive, bacterial, and gastro-intestinal issues, as well as for snake bites, asthma, and dizziness [ 68 , 76 , 77 ]. The bark and latex are intensely bitter, a reliable signal of the presence of bioactive secondary compounds and toxicity [ 94 – 96 ]. Budongo chimpanzees in both communities have been reported to consume both bark and dead wood of A . boonei , often travelling long distances to access these trees and only consuming small amounts of bark per feeding bout [ 45 ]. In an observation reported in this study (see Table 1 : A . boonei , Case 1 ), three males ingested A . boonei dead wood while outside the community’s core area for 1-minute. Two days before the event, one of the individuals had been observed with diarrhea, while also shedding visible tapeworm proglottids ( Bertiella sp.). This sample also contained unidentified protozoa, and Taenia sp. eggs. Pebsworth et al. [ 34 ] also reported an event in which four adult males, all with diverse parasite loads, traveled to a large A . boonei tree and ingested bark.
In the long-term site data, A . boonei bark ingestion was only documented 17 times between 2008–2021 [ 45 ], although this behavior was not systematically reported. In addition, the direct observation of only one A . boonei dead wood eating event, and no A . boonei bark ingesting events over the two four-month periods of observation in this study, suggest that consumption of this species is relatively rare across both communities. While specific pathogenic catalysts for selection of this species remain unknown, based on pharmacological, ethnobotanical, and behavioral data, we propose that A . boonei may be a therapeutic self-medicative resource for Budongo chimpanzees. The relatively strong inhibitory activity of this species against S . aureus , a bacteria associated with causing contamination on the skin leading to chronic wounds [ 97 ], as well as its anti-inflammatory properties, suggests that A . boonei ingestion may have beneficial effects in wound care contexts.
Khaya anthotheca . Previous studies have demonstrated that K . anthotheca bark contains biologically active compounds like gedunins, mexicanolide, phragmalin, and andirobins [ 98 ]. One limonoid identified in the species, anthothecol, has anti-cancer properties [ 99 ]. A study by Obbo et al. [ 100 ] on K . anthotheca bark collected in the Budongo Forest, found strong antiprotozoal activity against Plasmodium falciparum (IC 50 0.96 μg/mL) and Trypanosoma brucei rhodesiense (IC 50 5.72 μg/mL). A related species, K . senegalensis , has been shown to cause cell lysis in some gram-negative bacteria, including Salmonella Typhimurium , Escherichia coli , Shigella sp. and Salmonella sp., by targeting cytoplasmic membranes [ 101 ].
In our antibacterial library screen, of all extracts tested, only the methanol-water extract inhibited growth of A . baumannii (although no IC 50 values were reached in dose-response). The methanol-water extract also inhibited the growth of E . coli (DSM 498) in the library screen, as did the ethyl acetate (eE088) extract, though again no IC 50 values were reached. In our antibacterial dose-response assays, all extracts of K . anthotheca stem bark and resin exhibited strong inhibition against the Gram-positive E . faecium . The most active extract against this strain, which was also the strongest antibacterial result reported in this study, was methanol-water (mwE088) (IC 50 : 16 μg/mL; MIC: 32 μg/mL). All extracts of this species were also found to inhibit E . coli (DSM 1576) in the dose-response experiments, with the methanol-water extract once again also showing the strongest inhibition (IC 50 : 16 μg/mL; MIC: 256 μg/mL). This extract also inhibited the growth of S . maltophilia (IC 50 : 64 μg/mL) in the library screen. Only weak inhibition was found against the food pathogen S . enterica ( n -hexane extract, IC 50 : 256 μg/mL).
K . anthotheca exhibited potent anti-inflammatory activity. Of all extracts tested, the methanol-water K . anthotheca extract (mwE088) displayed the strongest COX-2 inhibition activity (IC 50 : 0.55 μg/mL). Past phytochemical studies on methanol and ethanol-water stem bark extracts from the related species, K . senegalensis , revealed many phenolic compounds, including flavonoids and tannins e.g., [ 101 , 102 ]. Flavonoids act on the inflammatory response, and may block molecules like COXs, cytokines, nuclear factor-кB and matrix metalloproteinases [ 103 ]. Some tannins have also been proven to have strong free radical-scavenging and antioxidant activities [ 104 ]. These compounds are antagonists of particular hormone receptors or inhibitors of particular enzymes such as COX enzymes [ 103 ]. If Khaya species are phytochemically similar, this could help explain K . anthotheca ’s strong COX-2 inhibitory activity.
Across Africa, K . anthotheca is traditionally used for ailments including allergies, fever, headaches, jaundice, bacterial infections, and as a disinfectant for bleeding wounds [ 105 – 107 ]. Our behavioral observations suggest that this species is also a common resource for Sonso chimpanzees, with a total of 65 feeding events recorded throughout the first field season. Of these events, several involved individuals with imbalanced health states (see Table 1 : K . anthotheca ) . On at least three independent occasions, K . anthotheca bark and resin were consumed by wounded individuals. Two adult females on different days tested positive for leukocytes on urinalysis tests within hours of ingesting K . anthotheca , suggesting the presence of infection. One of these individuals was also experiencing severe diarrhea the day prior, the other was found to have trace levels of blood in her urine. A juvenile female with a persistent cough was also observed consuming K . anthotheca bark. On several occasions individuals with high parasite loads or diverse species infection were observed targeting this resource while shedding tapeworm proglottids ( Bertiella sp.). An elderly female was also observed eating bark and resin a few hours prior to leaf-swallowing, a well-established self-medicative behavior known to rid the gut of endoparasites [ 9 , 23 ]. The frequency of K . anthotheca ingestion in the Sonso diet during this period, suggests that individuals have consistent exposure to the antibacterial and anti-inflammatory compounds present in this species. Whether this is a case of passive prevention through intake of a medicinal food, or therapeutic self-medication for a common and wide-spread condition will need further investigation. If used therapeutically, our results suggest this species could be used for treating wounds, bacterial or infections, and/or reducing internal parasite loads.
Christella parasitica.
Extracts of C . parasitica produced notably high anti-inflammatory activity in COX-2 testing, with the methanol-water extract (mwE087) achieving an IC 50 value of 0.81 μg/mL. This same extract, however, exhibited the lowest general activity in the antibacterial library screen. The only antibacterial activity from this species was on E . coli (DSM 498) by the ethyl acetate and n- hexane extracts (eE087; hE087), and on E . coli (DSM 1576) by the n-hexane extract (hE087). The n -hexane extract reached an IC 50 of 128 μg/mL in dose-response assays with no MIC value. Prior to this study, there had been limited pharmacological testing on C . parasitica (though see [ 108 ]), so comparison across studies is not possible.
When we considered the associated behavioral observation involving C . parasitica , we found a notable relevance to our pharmacological results (see Table 1 : C . parasitica , Case 1 ). This observation involved a wounded Sonso adult male (PS) travelling outside of his core area with a large group. It was unclear if this was an inter-community patrol. PS had been observed earlier in the day with a severe hand injury which impacted his mobility, though no open wound was observed. PS separated himself from the group and moved a few meters to a patch of ferns where he began consuming the leaflets. The bout lasted approximately 3-minutes. No other group members were observed feeding on this species, and this was only the second case of fern ingestion reported in Budongo in over 30-years of observations (unpublished site data). Health states of individuals from the past event were unfortunately not recorded. Whether or not C . parasitica ’s highly anti-inflammatory properties were the principal motivator for the selection of this species remains unknown, however, regardless of intention, this plant may have benefitted PS by reducing pain and swelling in his injured hand.
Syzygium guineense.
S . guineense bark and leaves have both previously been found to exhibit a range of pharmacological activity, reviewed by Uddin et al. [ 109 ]. The antioxidant, analgesic, and anti-inflammatory activities of this plant have been attributed to flavonoids, tannins, saponins, carbohydrates, alkaloids, and cardiac glycosides in the extracts [ 109 – 112 ]. In our assays, S . guineense bark exhibited high antibacterial growth inhibition effects in vitro . The methanol-water bark extract (mwE098a) showed some level of inhibition against all bacteria tested in the dose-response assays, except for E . faecium and S . enterica . This was also the only extract, out of all tested, to inhibit growth of P . aeruginosa (IC 50 : 64 μg/mL; MIC: >256 μg/mL) a pathogen known to cause infections in the blood, lungs, and other body parts after surgeries [ 113 ], and was one of two extracts to reach a MIC value against S . maltophilia (IC 50 : 32μg/mL; MIC: 256 μg/mL). The other extract to reach a MIC value was the ethyl acetate S . guineense bark extract (eE098a; IC 50 : 64 μg/mL; MIC: 256 μg/mL). All bark and leaf extracts showed strong inhibition against E . coli (DSM 1576) in the dose-response assays, with the strongest results coming from the methanol-water extracts (mwE098a and mwE098b). All bark and leaf extracts of this species, except for the n -hexane bark extract (hE098a), inhibited E . cloacae , and were the only extracts in the study to do so. E . cloacae , while part of normal intestinal flora, can cause UTI’s and respiratory infections in humans [ 114 ]. S . guineense extracts were also the only extracts to inhibit A . baumannii at a concentration <256 μg/mL, with the methanol-water bark extract showing the strongest inhibition. A . baumannii can cause infections in wounds, blood, urinary tracts, and lungs [ 115 ]. The efficacy of methanolic extracts from this species suggests that the active compounds are polar molecules. In the anti-inflammatory COX-2 inhibition dose-response assays, only the n -hexane bark extract displayed strong inhibitory effects (IC 50 : 2.42 μg/mL), while the other extracts failed to exhibit significant activity during the pre-screening or ≥ 50% inhibition at 10 μg/mL. The COX-2 inhibition assays showed no inflammatory inhibition amongst leaf extracts at tested concentrations.
S . guineense can be found throughout Sub-Saharan Africa and is a common traditional medicine, for malaria [ 116 ]. The bark is also used for stomach aches, diarrhea, internal parasites, and infertility [ 68 , 109 ]. Ingestion of S . guineense bark is rare in Budongo, with no direct observations in either community throughout the study period, and only six total cases between 2008–2021 documented in the site’s long-term data. No observations of leaf ingestion of this species have ever been reported. The infrequent ingestion of S . guineense bark implies a more targeted use, making it unlikely to be a medicinal food. Instead, our pharmacological findings make this resource a strong candidate as a putative, therapeutic self-medicative resource. Unfortunately, as there is currently no health data associated with individuals who have recently consumed S . guineense bark, we do not yet know which properties chimpanzees may be targeting. However, based on pharmacological results, we recommend further investigation into this species as a curative agent for respiratory-related infections.
Scutia myrtina.
Kritheka et al. [ 117 ] in their study on the bioactivity of S . myrtina , found in vivo evidence that this species possesses dose-dependent anti-inflammatory, antimicrobial, and antifungal properties. Across our antibacterial assays, the bark sample of this species collected from the stem inhibited E . faecium (eE089a) and E . coli DSM 1576 (eE089a; mwE089a) in dose-response tests at concentrations ≤256 μg/mL. The refuse sample, collected from the ground below the plant’s stem, inhibited A . baumannii (hE089b), E . faecium (eE089b), and E . coli DSM 1576 (mwE089b; eE089b; hE089b) in dose-response tests below the specified concentration. Interestingly, the refuse sample inhibited more bacteria species overall than the fresh bark. The most potent antibacterial growth inhibition effects came from the ethyl acetate bark sample against E . faecium (eE089a; IC 50 : 64 μg/mL), though no MIC value was reached. In the COX-2 inhibition assays, the n- hexane bark extract had the fifth strongest inhibitory effect in vitro (hE089a; IC 50 : 1.19 μg/mL) out of all samples, while the ethyl acetate refuse bark sample was less potent, though still moderately active (E089b; IC 50 : 7.49 μg/mL).
As far as the authors know, this is the first published report presenting both behavioral and pharmacological evidence for S . myrtina bark as a putative medicinal resource amongst free-ranging chimpanzees (though see [ 118 ] for evidence based on food-combinations). Our behavioral observations indicate that an individual with a diverse and intense parasite infection deliberately sought out the bark of this species. The Budongo chimpanzees may, therefore, utilize S . myrtina as an anthelminthic. Across traditional accounts from multiple regions, S . myrtina is commonly used by people as an anthelminthic to treat intestinal worms [ 68 ], while aerial parts are also used to treat various bacterial infections. As we were not able to conduct urinalysis on the consumer during or after this event, we cannot determine whether the individual also harbored a bacterial infection at the time of ingestion. However, this possibility cannot be ruled out. Based on these findings, we propose S . myrtina be added to the list of putative chimpanzee self-medication behaviors as a treatment for internal parasites, and we encourage further exploration into the other specific chimpanzee health conditions that this species may help ameliorate.
Assessment of putative self-medicative behaviors
We synthesized pharmacological and behavioral evidence to assess therapeutic use of species associated with bark feeding, dead wood eating, and pith stripping behaviors. A summary of the antibacterial and anti-inflammatory results for each species is reported in S3 Table in S2 File . Overall, stem bark and dead wood samples were notable for their activity. Bark samples from every species showed >40% antibacterial inhibition against at least one bacterial strain. This activity was also true of the dead wood samples. When plant parts of the same species were tested ( S . guineense and F . exasperata ), barks generally exhibited more potent antibacterial and COX-2 inhibition activity than the leaves, likely to do with the higher concentration of plant secondary metabolites in bark. Our findings offer strong support that bark and dead wood eating of certain species could constitute novel self-medicative behaviors in wild chimpanzees. We also encourage more investigation into the bioactivity of non-bitter pith stripping, as the pith of A . polystachius showed strong antibacterial activity against E . faecium (hE099; IC 50 : 32 μg/mL; MIC: 128 μg/mL), and the piths of both A . polystachius and M . leucantha demonstrated significant anti-inflammatory properties at low concentrations. Future primatological research should prioritize the establishment of multi-disciplinary long-term projects that look systematically at health states of individuals who engage in bark, dead wood, and pith ingestion behaviors. We also encourage further pharmacological testing on other species used for these behaviors in Budongo and across primate field sites.
Drug discovery
Multidisciplinary studies on this topic have potential to lead to the discovery of new medicines which may benefit our own species [ 119 – 122 ]. Historically, PSMs have played a major role in the development of modern human medicine, and even today, a large portion of medicines are derived either directly or indirectly from plants and other natural materials [ 123 – 127 ]. Antimicrobial resistance is rising to dangerously high levels according to the World Health Organization [ 128 ] requiring the rapid creation of new antibacterial treatments. Infections caused by multi-drug resistant bacteria kill hundreds of thousands of people annually. Our findings of strong antibacterial growth inhibition across numerous plant species growing in Budongo have promising implications for our ability to discover novel compounds in existing forest habitats. Extracts should also be tested against additional bacteria and for anti-virulence effects, e.g., inhibition and disruption of biofilm formation, quorum sensing and toxin production, pursuing development of new therapeutic strategies that apply less evolutionary pressure, likely resulting in emergence of less antibiotic resistances in the future. Phytochemical characterization using advanced techniques, such as LC-ToF-MS and NMR, as well as potentially AI-assisted untargeted metabolomics approaches, are now needed to identify substances present in the most active extracts. This may eventually lead to the isolation and structure elucidation of yet unknown active ingredients and make way for determining their pharmacological selectivity and toxicity, while also taking potential synergistic effects into account.
Simultaneously, we are currently faced with a pressing need for more effective treatments to combat symptoms of acute inflammation and mediate long-term consequences of chronic inflammatory diseases [ 129 ]. The prostaglandin-producing cyclooxygenase-2 (COX-2) mediates and regulates pain, fever, wound inflammation, and many other medical disorders, as it plays a crucial role in the host organism’s defense against pathogens and injury. COX-2 inhibition has the same mechanism of action as non-steroidal anti-inflammatory drugs (NSAIDs). While inflammation is a normal part of the body’s defense against injury or infection, it can be damaging when occurring in healthy tissues or over a protracted period. Chronic inflammation can lead to cardiovascular diseases (CVD) and cancer, the two leading global causes of death [ 130 ]. Past studies have shown that the IC 50 values of Aspirin and ibuprofen (pure compounds and common NSAIDs) are 210 μg/mL and 46 μg/mL respectively for COX-2, and 5 μg/mL and 1 μg/mL respectively for COX-1 [ 131 , 132 ]. The in vitro COX-2/COX-1 selectivity ratio for Aspirin and ibuprofen is 42 and 46 respectively. Surprisingly, the 17 most active extracts in our COX-2 assays display lower IC 50 values than these popular NSAIDs, meaning our extracts have more potent inhibitory effects on the inhibition of COX-2 than the most common anti-fever and anti-pain drugs on the market. While COX-1 assays were beyond the scope of this study, future research should investigate COX-1 inhibition activity of these 17 extracts to calculate COX-2/COX-1 selectivity ratios. Doing so will allow for preliminary assessment of potential side effects, selectivity, and efficacy before future in vivo experiments can commence.
Future directions
Future research on this topic would benefit from the inclusion of control samples (plants or plant parts not consumed by chimpanzees); however, in this study, assay costs were a prohibiting factor. Additional information regarding the nutritional and mineral content of the species mentioned in this study is needed to better understand the motivations for ingestion. However, bioactivity and nutritional/mineral content are by no means mutually exclusive. It is, therefore, highly likely that these resources provide multiple benefits to consumers.
Future studies should also consider ecological variables. For example, different individual plants of the same species should be tested across habitat types to determine whether bioactivity varies based on location, age, life history, or time of harvest. Situating samples in their ecological context will provide a better understanding of whether chimpanzees select resources based on species alone, or other more nuanced criteria. Lastly, climatic studies in combination with pharmacological testing should examine how climate change may impact bioactivity of these plants, as shifting weather patterns have already been shown to alter nutritional content [ 133 ]. This information will be critical for establishing protected habitats that can sustain healthy, wild, primate populations.
Conclusions
As we learn more about the pharmacological properties of plants ingested by chimpanzees in the wild, we can expand our understanding of their health maintenance strategies. Our results provide pharmacological evidence, from in vitro assays of plant parts consumed by wild chimpanzees collected in situ , for the presence of potent bioactive secondary plant metabolites in Budongo chimpanzee diets for a variety of potential illnesses previously not considered. Whether these resources are consumed intentionally as a form of therapeutic self-medication or passively as medicinal foods, must be assessed on a case-by-case basis, taking behavioral observations into account.
For the field of zoopharmacognosy to progress, we encourage continued multidisciplinary collaboration between primatologists, ethnopharmacologists, parasitologists, ecologists, and botanists [ 9 ]. Beyond improving our broad understanding of chimpanzee health maintenance, multidisciplinary studies will benefit our own species, potentially leading to the discovery of novel human medicines to combat the looming problem of growing drug-resistance. For this to happen, however, it is imperative that we urgently prioritize the preservation of our wild forest pharmacies as well as our primate cousins who inhabit them.
Materials availability
Voucher specimens for each species were deposited at the Makerere University Herbarium in Kampala, Uganda for taxonomic identification and storage. A duplicate set was deposited at the University of Oxford Herbarium for permanent storage.
Supporting information
S1 fig. budongo chimpanzees consuming resources tested in this study..
a.) IN eating K . anthotheca bark and resin b.) MZ eating S . myrtina bark c.) KC stripping A . polystachyus pith d.) MB eating C . patens dead wood e.) OZ eating S . guineense bark (post-study period) g.) MZ eating F . exasperata bark.
https://doi.org/10.1371/journal.pone.0305219.s001
S2 Fig. Generalized multi-method workflow used in this study.
https://doi.org/10.1371/journal.pone.0305219.s002
S3 Fig. Voucher samples collected in duplicate.
a . ) C . alexandri (00243133G) b . ) A . polystachius (00243136J) c . ) W . elongata (00243129L) d . ) C . parasitica (00243122E) e . ) K . anthotheca (00243123F) f . ) F . variifolia (51195) g . ) M . leucantha (51203) h . ) A . boonei (51204) i . ) D . dewevrei (00243132F) j . ) S . guineense (00243135I) k . ) S . myrtina (00243128K) l . ) F . exasperata (00243130D).
https://doi.org/10.1371/journal.pone.0305219.s003
S4 Fig. Plate layouts for growth inhibition assays.
[Top] Library Screen: done in 96-wells-mikrotiterplate; AB: Antibiotic as positive control; DMSO: vehicle control / negative control; GC: growth control: containing working culture, to check whether the bacterium grew/active; [Bottom] Dose-Response: done in descending concentration of samples, DMSO, and antibiotic. MB: Media blank, consisted of CAMHB as negative/ sterile media control; DMSO as negative/ vehicle control; GC: growth control, consisted of working culture.
https://doi.org/10.1371/journal.pone.0305219.s004
S5 Fig. ELISA assay setup for anti-inflammatory assay.
https://doi.org/10.1371/journal.pone.0305219.s005
S1 File. Supplementary materials: Methods .
https://doi.org/10.1371/journal.pone.0305219.s006
S2 File. Supplementary tables.
https://doi.org/10.1371/journal.pone.0305219.s007
Acknowledgments
We are grateful to all the field staff working in Budongo who provided invaluable instruction and guidance, generously sharing both scientific insight and traditional knowledge. This study could not have been done without their contributions. Specifically, we would like to thank members of the Perspectives Collective: Chandia Bosco, Monday Mbotella Gideon, Adue Sam, Asua Jackson, Steven Mugisha, Atayo Gideon, and Kizza Vincent, and Walter Akankwasa, as well as site director David Eryenyu. We would also like to thank Godwin Anywar for his assistance with plant identification at the Makerere Herbarium, Stephen Harris at the University of Oxford’s Herbarium for his facilitation of voucher storage, and the Natural History Museum in London for their aid in parasite identification. We are grateful to Vernon Reynolds who founded the field site and to the Royal Zoological Society of Scotland for providing core support. We also gratefully acknowledge the Uganda Wildlife Authority and the Uganda National Council for Science and Technology for granting permission to conduct research in Uganda. Lastly, thank you to the staff and students at Neubrandenburg University of Applied Sciences who made this collaboration possible, and to research assistant, Finn Freymann, for his help with botanical extractions.
- View Article
- PubMed/NCBI
- Google Scholar
- 7. Toft CA, Aeschlimann A, Bolis L. Parasite-host associations: Coexistence or conflict? Oxford University Press (OUP); 1991.
- 12. MacIntosh AJJ, Huffman MA. Topic 3: Toward Understanding the Role of Diet in Host–Parasite Interactions: The Case for Japanese Macaques BT—The Japanese Macaques. In: Nakagawa N, Nakamichi M, Sugiura H, editors. Tokyo: Springer Japan; 2010. pp. 323–344. https://doi.org/10.1007/978-4-431-53886-8_15
- 19. Wrangham RW. Behavioural ecology of chimpanzees in Gombe National Park, Tanzania. University of Cambridge; 1975.
- 42. Russak S. Ecological role of dry-habitat chimpanzees (Pan troglodytes schweinfurthii) at Issa, Ugalla, Tanzania. Arizona State University. 2013. Available: https://repository.asu.edu/items/18012
- 43. Matsuzawa T, Humle T, Sugiyama Y. The chimpanzees of Bossou and Nimba. Springer; 2011.
- 55. Reynolds LBAFV, Reynolds V, Goodall J, Press OU. The Chimpanzees of the Budongo Forest: Ecology, Behaviour and Conservation. OUP Oxford; 2005. Available: https://books.google.co.uk/books?id=NnwSDAAAQBAJ
- 61. Lozano GA. Parasitic Stress and Self-Medication in Wild Animals. In: Møller AP, Milinski M, Slater PJBBT-A in the S of B, editors. Stress and Behavior. Academic Press; 1998. pp. 291–317. https://doi.org/10.1016/S0065-3454(08)60367-8
- 62. The World Health Organization (WHO). Bench aids for the diagnosis of intestinal parasites, second edition. Geneve; 2019.
- 68. Kokwaro JO. Medicinal plants of East Africa. Kampala: University of Nairobi Press; 1976. Available: https://books.google.co.uk/books?id=msyHLY0dhPwC
- 71. Akhondi H. Bacterial Diarrhea. Simonsen KA, editor. 2022. Available: https://www.ncbi.nlm.nih.gov/books/NBK551643/#_NBK551643_pubdet_
- 72. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI supplement M100. 30th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- 77. Burkill HM. Dalziel JM, Hutchinson J. The useful plants of west tropical Africa. 2nd ed. The useful plants of west tropical Africa, Vols. 1–3. Royal Botanic Gardens, Kew; 1995.
- 81. Terashima H, Kalala S, Malasi N. Ethnobotany of the Lega in the tropical rain forest of Eastern Zaire. African study monographs. Center for African Area Studies, Kyoto University; 1991.
- 82. Howard P, Butono F, Kayondo-Jjemba P, Muhumuza C. Integrating forest conservation into district development: A case study. In P. Howard (ed.), Na- ture conservation in Uganda’s tropical forest reserves. Glanda, Switzerland, and Cambridge, U.K.; 1991.
- 83. PROTA. PROTA4U. 2023. Available: https://prota.prota4u.org/
- 95. Crellin JK, Philpott J, Bass ALT. Herbal Medicine Past and Present: A reference guide to medicinal plants. Duke University Press; 1990.
- 96. Githens TS. Drug plants of Africa. University of Pennsylvania Press; 2017.
- 105. Akoègninou A, Van der Burg WJ, Van der Maesen LJG. Flore analytique du Bénin. Backhuys Publishers; 2006.
- 119. Huffman MA H. O, Kawanaka M, Page JE, Kirby GC, Gasquet M, et al. African great ape self-medication: A new paradigm for treating parasite disease with natural medicines? In: Ebizuka Y, editor. Towards Natural Medicine Research in the 21st Century. Amsterdam: Elsevier Science B.V.; 1998. pp. 113–123.
- 122. Rodriguez E, Wrangham R. Zoopharmacognosy: The Use of Medicinal Plants by Animals. In: Downum KR, Romeo JT, Stafford HA, editors. Phytochemical Potential of Tropical Plants. Boston, MA: Springer US; 1993. pp. 89–105. https://doi.org/10.1007/978-1-4899-1783-6_4
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Dissertation
- How to Write a Discussion Section | Tips & Examples
How to Write a Discussion Section | Tips & Examples
Published on 21 August 2022 by Shona McCombes . Revised on 25 October 2022.

The discussion section is where you delve into the meaning, importance, and relevance of your results .
It should focus on explaining and evaluating what you found, showing how it relates to your literature review , and making an argument in support of your overall conclusion . It should not be a second results section .
There are different ways to write this section, but you can focus your writing around these key elements:
- Summary: A brief recap of your key results
- Interpretations: What do your results mean?
- Implications: Why do your results matter?
- Limitations: What can’t your results tell us?
- Recommendations: Avenues for further studies or analyses
Instantly correct all language mistakes in your text
Be assured that you'll submit flawless writing. Upload your document to correct all your mistakes.

Table of contents
What not to include in your discussion section, step 1: summarise your key findings, step 2: give your interpretations, step 3: discuss the implications, step 4: acknowledge the limitations, step 5: share your recommendations, discussion section example.
There are a few common mistakes to avoid when writing the discussion section of your paper.
- Don’t introduce new results: You should only discuss the data that you have already reported in your results section .
- Don’t make inflated claims: Avoid overinterpretation and speculation that isn’t directly supported by your data.
- Don’t undermine your research: The discussion of limitations should aim to strengthen your credibility, not emphasise weaknesses or failures.
Prevent plagiarism, run a free check.
Start this section by reiterating your research problem and concisely summarising your major findings. Don’t just repeat all the data you have already reported – aim for a clear statement of the overall result that directly answers your main research question . This should be no more than one paragraph.
Many students struggle with the differences between a discussion section and a results section . The crux of the matter is that your results sections should present your results, and your discussion section should subjectively evaluate them. Try not to blend elements of these two sections, in order to keep your paper sharp.
- The results indicate that …
- The study demonstrates a correlation between …
- This analysis supports the theory that …
- The data suggest that …
The meaning of your results may seem obvious to you, but it’s important to spell out their significance for your reader, showing exactly how they answer your research question.
The form of your interpretations will depend on the type of research, but some typical approaches to interpreting the data include:
- Identifying correlations , patterns, and relationships among the data
- Discussing whether the results met your expectations or supported your hypotheses
- Contextualising your findings within previous research and theory
- Explaining unexpected results and evaluating their significance
- Considering possible alternative explanations and making an argument for your position
You can organise your discussion around key themes, hypotheses, or research questions, following the same structure as your results section. Alternatively, you can also begin by highlighting the most significant or unexpected results.
- In line with the hypothesis …
- Contrary to the hypothesised association …
- The results contradict the claims of Smith (2007) that …
- The results might suggest that x . However, based on the findings of similar studies, a more plausible explanation is x .
As well as giving your own interpretations, make sure to relate your results back to the scholarly work that you surveyed in the literature review . The discussion should show how your findings fit with existing knowledge, what new insights they contribute, and what consequences they have for theory or practice.
Ask yourself these questions:
- Do your results support or challenge existing theories? If they support existing theories, what new information do they contribute? If they challenge existing theories, why do you think that is?
- Are there any practical implications?
Your overall aim is to show the reader exactly what your research has contributed, and why they should care.
- These results build on existing evidence of …
- The results do not fit with the theory that …
- The experiment provides a new insight into the relationship between …
- These results should be taken into account when considering how to …
- The data contribute a clearer understanding of …
- While previous research has focused on x , these results demonstrate that y .
Even the best research has its limitations. Acknowledging these is important to demonstrate your credibility. Limitations aren’t about listing your errors, but about providing an accurate picture of what can and cannot be concluded from your study.
Limitations might be due to your overall research design, specific methodological choices , or unanticipated obstacles that emerged during your research process.
Here are a few common possibilities:
- If your sample size was small or limited to a specific group of people, explain how generalisability is limited.
- If you encountered problems when gathering or analysing data, explain how these influenced the results.
- If there are potential confounding variables that you were unable to control, acknowledge the effect these may have had.
After noting the limitations, you can reiterate why the results are nonetheless valid for the purpose of answering your research question.
- The generalisability of the results is limited by …
- The reliability of these data is impacted by …
- Due to the lack of data on x , the results cannot confirm …
- The methodological choices were constrained by …
- It is beyond the scope of this study to …
Based on the discussion of your results, you can make recommendations for practical implementation or further research. Sometimes, the recommendations are saved for the conclusion .
Suggestions for further research can lead directly from the limitations. Don’t just state that more studies should be done – give concrete ideas for how future work can build on areas that your own research was unable to address.
- Further research is needed to establish …
- Future studies should take into account …
- Avenues for future research include …

Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
McCombes, S. (2022, October 25). How to Write a Discussion Section | Tips & Examples. Scribbr. Retrieved 24 June 2024, from https://www.scribbr.co.uk/thesis-dissertation/discussion/
Is this article helpful?
Shona McCombes
Other students also liked, how to write a results section | tips & examples, research paper appendix | example & templates, how to write a thesis or dissertation introduction.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 18 June 2024
Plasma proteomics identify biomarkers predicting Parkinson’s disease up to 7 years before symptom onset
- Jenny Hällqvist ORCID: orcid.org/0000-0001-6709-3211 1 , 2 na1 ,
- Michael Bartl ORCID: orcid.org/0000-0002-7752-2443 3 , 4 na1 ,
- Mohammed Dakna 3 ,
- Sebastian Schade ORCID: orcid.org/0000-0002-6316-6804 5 ,
- Paolo Garagnani ORCID: orcid.org/0000-0002-4161-3626 6 ,
- Maria-Giulia Bacalini 7 ,
- Chiara Pirazzini 6 ,
- Kailash Bhatia ORCID: orcid.org/0000-0001-8185-286X 8 ,
- Sebastian Schreglmann ORCID: orcid.org/0000-0002-4129-5808 8 ,
- Mary Xylaki ORCID: orcid.org/0000-0002-7892-8621 3 ,
- Sandrina Weber 3 ,
- Marielle Ernst 9 ,
- Maria-Lucia Muntean 5 ,
- Friederike Sixel-Döring 5 , 10 ,
- Claudio Franceschi ORCID: orcid.org/0000-0001-9841-6386 6 ,
- Ivan Doykov 1 ,
- Justyna Śpiewak 1 ,
- Héloїse Vinette ORCID: orcid.org/0009-0000-4360-1293 1 , 11 ,
- Claudia Trenkwalder 5 , 12 ,
- Wendy E. Heywood ORCID: orcid.org/0000-0003-2106-8760 1 ,
- Kevin Mills 2 na2 &
- Brit Mollenhauer ORCID: orcid.org/0000-0001-8437-3645 3 , 5 na2
Nature Communications volume 15 , Article number: 4759 ( 2024 ) Cite this article
28k Accesses
2173 Altmetric
Metrics details
- Parkinson's disease
Parkinson’s disease is increasingly prevalent. It progresses from the pre-motor stage (characterised by non-motor symptoms like REM sleep behaviour disorder), to the disabling motor stage. We need objective biomarkers for early/pre-motor disease stages to be able to intervene and slow the underlying neurodegenerative process. Here, we validate a targeted multiplexed mass spectrometry assay for blood samples from recently diagnosed motor Parkinson’s patients ( n = 99), pre-motor individuals with isolated REM sleep behaviour disorder (two cohorts: n = 18 and n = 54 longitudinally), and healthy controls ( n = 36). Our machine-learning model accurately identifies all Parkinson patients and classifies 79% of the pre-motor individuals up to 7 years before motor onset by analysing the expression of eight proteins—Granulin precursor, Mannan-binding-lectin-serine-peptidase-2, Endoplasmatic-reticulum-chaperone-BiP, Prostaglaindin-H2-D-isomaerase, Interceullular-adhesion-molecule-1, Complement C3, Dickkopf-WNT-signalling pathway-inhibitor-3, and Plasma-protease-C1-inhibitor. Many of these biomarkers correlate with symptom severity. This specific blood panel indicates molecular events in early stages and could help identify at-risk participants for clinical trials aimed at slowing/preventing motor Parkinson’s disease.
Similar content being viewed by others

Meta-analysis of shotgun sequencing of gut microbiota in Parkinson’s disease

The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins
Target engagement and immunogenicity of an active immunotherapeutic targeting pathological α-synuclein: a phase 1 placebo-controlled trial
Introduction.
Parkinson’s disease (PD) is a complex and increasingly prevalent neurodegenerative disease of the central nervous system (CNS). It is clinically characterised by progressive motor and non-motor symptoms that are caused by α-synuclein aggregation predominantly in dopaminergic cells, which leads to Lewy body (LB) formation 1 . The failure of neuroprotective strategies in preventing disease progression is due, in part, to the clinical heterogeneity of the disease—it has several phenotypes—and to the lack of objective biomarker readouts 2 . To facilitate the approval of neuroprotective strategies, governing agencies and pharmaceutical companies need regulatory pathways that use objectively measurable markers—potential therapeutical targets as well as state and rate biomarkers—directly associated with PD pathophysiology and clinical phenotypes 3 .
The recently emerged α-synuclein seed amplification assays (SAA) can identify α-synuclein pathology in vivo and support stratification purposes but still rely on cerebrospinal fluid (CSF) obtained through relatively invasive lumbar punctures 4 . Therefore, this test remains specialised and not readily suitable for large-scale clinical use. As peripheral fluid biomarkers are less invasive and easier to obtain, they could be used in repeated and long-term monitoring, which is necessary for population-based screenings for upcoming neuroprotective trials. While the only emerged serum biomarker in the last years, axonal marker neurofilament light chain (NfL), increases longitudinally and correlates with motor and cognitive PD progression 5 , it is non-specific to the disease process.
Growing data support evidence of PD pathology in the peripheral system, which increases the likelihood of finding a source of matrices for less invasive biomarkers. We know α-synuclein aggregation induces neurodegeneration, which is propagated throughout the CNS. Evidence indicates that additional inflammatory events are an early and potentially initial step in a pathophysiological cascade leading to downstream α-synuclein aggregation that activates the immune system 6 . Inflammatory risk factors in circulating blood (i.e. C-reactive-protein and Interleukin-6 and α-synuclein-specific T-cells) are associated with motor deterioration and cognitive decline in PD 7 , 8 . These inflammatory blood markers can even be identified in plasma/serum samples of individuals with isolated REM sleep behaviour disorder (iRBD), the early stage of a neuronal synuclein disease (NSD), and the most specific predictor for PD and dementia with Lewy bodies (DLB) 6 . NSD was recently proposed as a biologically defined term, for a spectrum of clinical syndromes, including iRBD, PD and DLB, that follow an integrated clinical staging system of progressing neuronal α-synuclein pathology (NSD-ISS) 9 .
In this study, we used mass spectrometry-based proteomic phenotyping to identify a panel of blood biomarkers in early PD. In the initial discovery stage, we analysed samples from a well-characterised cohort of de novo PD patients and healthy controls (HC) who had been subjected to rigorous collection protocols 10 . Using unbiased state-of-the-art mass spectrometry, we identified putatively involved proteins, suggesting an early inflammatory profile in plasma. We thereafter moved on to the validation phase by creating a high-throughput and targeted proteomic assay that was applied to samples from an independent replication cohort, consisting of de novo PD, HC and iRBD patients. Finally, after refining the targeted proteomic panel to include a multiplex of only the biomarkers which were reliably measured, an independent analysis was performed on a larger and independent cohort of longitudinal, high-risk subjects who had been confirmed as iRBD by state-of-the-art video-recorded polysomnography (vPSG), including follow-up sampling of up to 7 years.
In summary, using a panel of eight blood biomarkers identified in a machine-learning approach, we were able to differentiate between PD and HC with a specificity of 100%, and to identify 79% of the iRBD subjects, up to 7 years before the development of either DLB or motor PD (NSD stage 3). Our identified panel of biomarkers significantly advances NSD research by providing potential screening and detection markers for use in the earliest stages of NSD for subject identification/stratification for the upcoming prevention trials.
Proteomic discovery phase 0
We performed a bottom-up proteomics analysis of plasma, which had been depleted of the major blood proteins, using two-dimensional in-line liquid chromatography fractionation into ten fractions and label-free mass spectrometric analysis by QTOF MS E . The discovery cohort consisted of ten randomly selected drug-naïve patients with PD and ten matched HC from the de novo Parkinson’s disease (DeNoPa 10 ) cohort (details can be found in Supplementary Table 1 ). This analysis identified 1238 proteins when restricting identification to originate from at least one peptide per protein and at least two fragments per peptide. After excluding proteins with less than two unique peptides or with an identification score below a set threshold (see method section below), 895 distinct proteins remained. Of these proteins, 47 were differentially expressed between the de novo PD and control groups on a nominal significance level of 95%. Pathway analysis suggested enrichment in several inflammatory pathways. Workflow and Results are shown in Fig. 1 , and 2 Supplementary Figs. 1 , 2.

The study included three phases. Phase 0 consisted of discovery proteomics by untargeted mass spectrometry to identify putative biomarkers, followed by phase I in which targets from the discovery phase were transferred to a targeted, mass spectrometric MRM method and applied to a new and larger cohort of samples, and finally phase II in which the targeted MRM method was refined and a larger number of samples were analysed to evaluate the clinical feasibility of the targeted protein panel.
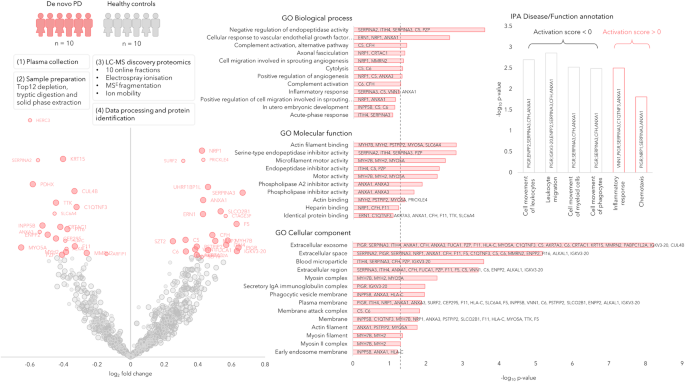
The circle radii in the Volcano plot represent the identification certainty, where large radii represent proteins identified by at least two unique peptides and an identification score >15, smaller radii are given for proteins identified by two or more unique peptides or a confidence score >15. The horizontal axis shows log 2 of the average fold-change and the vertical axis shows −log 10 of the p values. The significantly different proteins are annotated by gene name and coloured in pink, while the non-significant proteins are coloured in grey. GO annotations for the significant proteins are shown, the dashed line represents p = 0.05. Disease and function annotations from IPA are shown, divided into annotations with a positive or negative activation score. Source data are provided as a Source Data file.
Selection of proteins for the targeted proteomic assay
We next developed a validatory, high-throughput and multiplexed, mass spectrometric targeted proteomic assay based on the potential biomarkers identified in the discovery phase. Additional proteins were also included in the assay, several of which had been identified in previous discovery studies of PD, Alzheimer’s disease (AD), and ageing 11 . In addition, we also included several known pro- and anti-inflammatory proteins identified in the literature 12 , 13 , 14 , 15 , which had been previously developed into an in-house targeted proteomic neuroinflammatory panel. Using this approach, we created a targeted proteomic panel, including biomarkers from current scientific developments and preliminary findings from our own work 16 , 17 . This targeted proteomic and multiplexed assay included 121 proteins and aimed to validate biomarkers and probe the pathways identified as being perturbed in the discovery phase. Details can be found in Supplementary Table 2 and Fig. 3 .
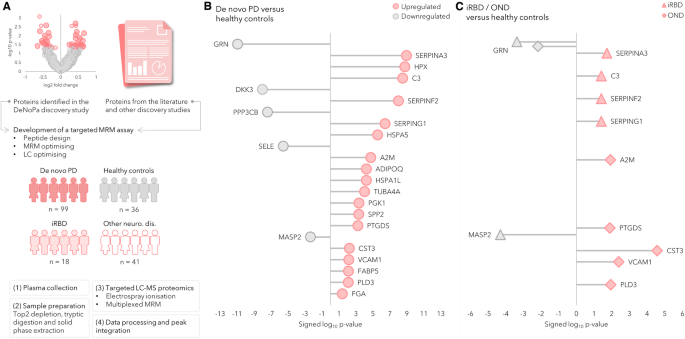
Workflow and overview of the results of the targeted proteomic analysis of de novo Parkinson’s disease (PD) subjects, healthy controls (HC), and the validation cohorts of other neurological disorders (OND) and isolated REM sleep behaviour disorder (iRBD). A A targeted mass spectrometric proteomic assay was developed and optimised. The assay was then applied to plasma samples from cohorts comprising de novo PD ( n = 99) and HC ( n = 36), and validated in patients with OND ( n = 41) and prodromal subjects with iRBD ( n = 18). The protein expression difference between the groups was compared using Mann–Whitney’s two-sided U -test with Benjamini–Hochberg FDR adjustment at 5%. The lollipop charts show the log 10 p values, signed according to fold-changes. Pink icons represent a protein upregulated in an affected group and grey represents a protein upregulated in controls. B Significantly differentially expressed proteins in the comparison between de novo PD and healthy controls. C Significantly differentially expressed proteins between iRBD, OND and HC. Source data are provided as a Source Data file.
Demographics-targeted proteomic validation phase (phase I)
For the targeted proteomics analysis, we used plasma samples, independent from the proteomic discovery step, from 99 individuals recently diagnosed with de novo PD (48 men, 50%, mean age 67 years) and 36 healthy controls (HC; 20 men, 57%, mean age 64 years). This was the main cohort, to which we added further samples for validation that consisted of a heterogeneous group of 41 patients with other neurological diseases (OND) (29 men, 71%, mean age 70 years) and 18 patients with vPSG-confirmed iRBD (10 men, 56%, mean age 67 years). Further details can be found in Table 1 and Fig. 3 .
The identification of biomarkers that were significantly and differentially expressed biomarkers between patients with de novo Parkinson’s disease and healthy controls- Targeted proteomic validation phase (phase I)
Our targeted proteomic assay was developed for 121 proteins, 32 of which we consistently and reliably detected in plasma. Of these 32 markers, 23 were confirmed as being significantly and differentially expressed between PD and HC. We identified six differentially expressed proteins in the comparison between iRBD patients and HC and between OND and HC (Fig. 3 ). Both the de novo PD and iRBD groups demonstrated an upregulated expression of the serine protease inhibitors SERPINA3, SERPINF2 and SERPING1, and of the central complement protein C3. Granulin precursor protein was shown to be downregulated in all three patient groups (PD, iRBD and OND) compared to HC. The OND and PD groups had a shared and upregulated expression of the proteins PTGDS, CST3, VCAM1 and PLD3. Detailed information about the diagnoses of the OND group can be found in Table 1 , and detailed information about the proteins can be found in Supplementary Table 2 . Figure 4 shows the significantly different proteins as Box-scatter plots.

The data are displayed as Box and Whisker plots overlaid with scatter plots of the individual measurements. The whiskers show the minimum and maximum, and the boxes show the 25th percentile, the median and the 75th percentile. The protein expression difference between the groups was compared using Mann–Whitney’s two-sided U -test with Benjamini–Hochberg multiple testing correction (FDR adjustment at 5%). ns not significant, * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001. The proteins are represented by gene names. Source data are provided as a Source Data file.
The biological significance of the differentially expressed proteins- Targeted proteomic validation phase (phase I)
The involvement of the differentially expressed proteins and their impact on biological processes were evaluated using pathway analysis (Ingenuity Pathway Analysis [IPA], Qiagen). The significantly differentially expressed proteins between PD and HC were used as input, with a fold-change set as the expression observation. We considered pathways as significant if they had an enrichment p value <0.05. At least two of the input proteins were included. Three major pathway clusters were identified and consisted of (i) the expression of serine protease inhibitors or serpins and complement and coagulation components, (ii) endoplasmic reticulum (ER) stress/heat shock-related proteins and (iii) the expression of VCAM1, SELE and PPP3CB. The highest enrichment scores were identified in the pathways acute phase response signalling ( p = 7.8 E −10 ), coagulation system ( p = 7.4 E −6 ), complement system ( p = 8.1 E −6 ), LXR/RXR activation ( p = 9.1 E −6 ), FXR/RXR activation ( p = 9.8 E −6 ) and glucocorticoid receptor signalling ( p = 2.0 E −5 ). These are all pathways involved in inflammatory responses. We also identified pathways related to the unfolded protein response ( p = 0.004) and neuroinflammation ( p = 0.04), although with lower enrichment scores. For details, see Supplementary Fig. 1 .
Inflammation-related pathways (including both the complement system and the acute phase response) demonstrated the highest significance levels, followed by pathways regulating protein folding, ER stress, and heat shock proteins. A network representation of proteins and pathways showed clusters consisting of inflammation/coagulation/lipid metabolism (FXR/RXR and LXR/RXR), heat shock proteins/protein misfolding, and more heterogenous pathway clusters related to Wnt-signalling and extracellular matrix proteins. Figure 5 illustrates the potential detrimental and protective mechanisms suggested to be taking place based on the protein expressions observed in this study, leading to oligomerisation and accumulation of α-synuclein in neuronal Lewy body inclusions and, finally, dopaminergic neuronal cell loss.

Oligomerisation and accumulation of α-synuclein in Lewy body inclusions is a key process in the pathophysiology of neuronal synuclein disease, i.e. Parkinson’s disease and dementia with Lewy bodies from aggregation and accumulation, the pathological pathway includes different steps finally leading to the loss of dopaminergic neurons. Protective and detrimental mechanisms influence these processes, based on the differently expressed protein profiles, assessed by targeted mass spectrometry in our study. Detailed information about the proteins can be found in Supplementary Table 2 .
Multivariate analysis shows differences between the proteomes of Parkinson’s disease and controls- Targeted proteomic validation phase (phase I)
Principal component analysis (PCA) demonstrated that the HC and PD groups formed two clusters separate from each other over the first and second principal components (PC), attributed with 23.5% and 13.9% of the model’s total variance, respectively. The iRBD group was situated in the middle of HC and PD, and the OND group varied considerably with no evident clustering, as expected due to the heterogeneity of diseases. The corresponding loadings of PC1 and PC2 demonstrated that those with PD correlated with lower levels of PPP3CB, DKK3, SELE and GRN, and higher levels of most of the other proteins. The loadings plot had a high level of covariation in the expression of the PPP3CB, DKK3 and SELE proteins, which were all downregulated in PD. These proteins correlated negatively with the expression of SERPINs, complement C3 and HPX, which all showed a high degree of covariation, and were upregulated in the PD group. Data are displayed in Supplementary Fig. 2 .
The use of multiplexed protein panels of protein biomarkers for the prediction of de novo Parkinson’s disease- Targeted proteomic validation phase (phase I)
We next applied machine learning to construct a discriminant OPLS-DA model using the PD and HC samples from the validation phase. The samples clustered into two distinct and well-separated classes, and evaluation of the model showed that it was highly significant ( p = 2.3E −27 permutations p = <0.001). The proteins with the greatest influence on the class separations were GRN, DKK3, C3, SERPINA3, HPX, SERPINF2, CAPN2, SERPING1 and SELE. We predicted the iRBD samples in the model, which resulted in 13 subjects classified as PD (72%) and five not belonging to either group. None of the iRBD samples were classified as controls. We additionally predicted the OND samples, out of which nine were classified as HC, 12 as PD and 19 were not classified as belonging to either group. The 12 samples predicted as PD did not demonstrate enrichment according to the OND groups. The random distribution of the OND samples between PD and HC indicates that the heterogenous group of OND individuals does not share a distinct protein expression with either the HC or PD groups. The iRBD samples that were classified as PD, and not as HC, strongly suggest a shared proteomic profile between iRBD and the protein expression observed in the newly diagnosed PD patients.
We subsequently explored if the observed protein expressions could be used to build a regression model capable of predicting whether individuals belonged to the PD or HC groups. We identified a panel of proteins that discriminated between PD and HC with 100% accuracy and then constructed a linear support vector classification model and applied recursive feature elimination to pinpoint the most discriminating variables. The data were divided into two parts: one consisting of 70% for model training and one containing 30% for testing. The proportion of PD and control samples was maintained in each part. The number of features included in the model was determined by feature ranking with cross-validated recursive feature elimination in the training dataset. The feature selection resulted in a model with eight predictors: GRN, MASP2, HSPA5, PTGDS, ICAM1, C3, DKK3 and SERPING1. The training data were predicted in the model and resulted in all samples being classified in the correct class. We further constructed receiver operating characteristic (ROC) and precision-recall (PR) curves to illustrate the ability of each protein to distinguish between PD and HC and compared this with the ability of the combined multiplexed protein panel. The combined panel achieved an AUC of 1.0 on both ROC and PR curves. The AUC of the individual predictors ranged from 0.53 to 0.92 in the ROC curve, and from 0.79 to 0.96 in the PR curve (Fig. 6 ). We further evaluated the whole dataset by performing repeated cross-validation with six splits of the data and 40 repetitions. The resulting classification metrics (Supplementary Fig. 3 ) demonstrated average and standard deviation for precision, recall, F1 score, and balanced accuracy score of 0.87 ± 0.09, 0.87 ± 0.08, 0.86 ± 0.09 and 0.82 ± 0.12, respectively, thereby indicating a highly robust classification model. Testing the model’s specificity for PD, we predicted the heterogenous group of OND, resulting in 26 of the 42 samples being classified as PD-like. Prediction of the prodromal iRBD group resulted in 17 of 18 samples being classified as PD-like. We compared the prediction of the OND and iRBD samples between the OPLS-DA and SVM models, finding that most of the samples were classified in the same group in both models (out of the samples with a classification in the OPLS-DA model: 82% in OND and 100% in iRBD). The proportion of iRBD samples classified as PD in our models (72% in the OPLS-DA model and 94% in the SVM model) is in line with clinical evidence based on longitudinal cohort studies, reporting that over 80% of iRBD subjects will develop an advanced NSD with motor impairment and/or cognitive decline 18 . We evaluated the influence of age and sex on the proteins included in the support vector model and found that neither influenced the model’s classification ability (see Supplementary Methods 2 for details).

The model was trained on 70% of the samples to establish the most discriminating features. Applying cross-validated recursive feature elimination, the top predictors were determined as a granulin precursor, mannan-binding lectin-serine peptidase 2, endoplasmic reticulum chaperone-BiP, prostaglandin-H2 d -isomerase, intercellular adhesion molecule-1, complement C3, dickkopf-3 and plasma protease C1 inhibitor. The remaining 30% of samples were predicted in the model and resulted in 100% prediction accuracy. Receiver operating characteristics (ROC) and precision-recall (PR) curves of the individual and combined proteins in the test set demonstrated that the individual proteins achieved ROC area under the curve (AUC) values 0.53–0.92 and PR values 0.79–0.96, while the combined predictors reached an area under the curve = 1.0. Source data are provided as a Source Data file.
Development of a rapid and refined LC-MS/MS method and evaluation of an independent and longitudinal iRBD cohort (Independent replication cohort-phase II)
To evaluate the results from the initial prediction models focusing on at-risk subjects, we developed and refined our targeted and multiplexed proteomic test to quantitate only those proteins that were readily and reliably detectable from the initial targeted proteomic assay ( n = 32). Next, we analysed an additional set of 146 longitudinal samples from an independent cohort of 54 individuals with iRBD. This cohort was available from continuing recruitment at the same centre and consisted of longitudinally followed iRBD subjects. Deep phenotyping revealed 100% (54/54) had RBD on PSG, 88.9% (48/54) had hyposmia as identified with the Sniffin’ Stick Identification Test, and 91.7 % (22/24) had neuronal α-synuclein positivity as shown by α-synuclein Seed Amplification Assay (SAA) in cerebrospinal fluid (CSF) 19 . Longitudinal follow-up was available for up to 10 years, during which 16 subjects (20%) phenoconverted to either PD ( n = 11) or dementia with Lewy bodies (DLB; n = 5). Since only serum samples were available from the independent replication cohort (further details can be found in Supplementary Table 3 ), we investigated how the proteins in our assay correlated between plasma, serum, and CSF and found good correlations between plasma and serum, but poor correlations between these blood matrices and CSF. The limited correlations between blood and CSF proteins correspond to those of other studies comparing the protein expression between plasma/serum and CSF 20 , 21 and underscore that our test does not necessarily reflect a prodromal and PD-specific proteomic signature of the protein expression in the CSF in proximity to the brain, but rather shows an earlier change in the blood protein expression between healthy status and very early PD patients (Details from this comparison can be found in Supplementary Methods 1 and Supplementary Fig. 4 ).
We applied all available longitudinal iRBD samples ( n = 146) from phase II to the two machine-learning models (OPLS-DA and support vector machine) constructed in phase I (PD vs. HC). The OPLS-DA model, based on all 32 detected proteins, identified 70% of the iRBD samples as PD, while the SVM model, which was based on a panel of eight proteins, identified 79% of the samples as PD. As mentioned above, at the time of analysis, 16 of the 54 subjects in our longitudinal iRBD validation cohort had developed PD/DLB. The earliest correct classification was 7.3 years prior to phenoconversion and the latest was 0.9 years prior to diagnosis (average 3.5 ± 2.4 years). Detailed information can be found in Fig. 7 and Supplementary Methods 3 .
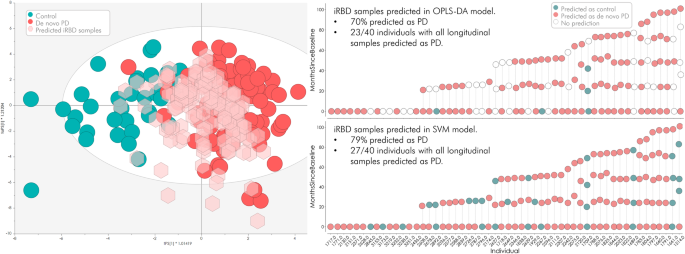
146 new serum samples from individuals diagnosed with iRBD, several with longitudinal follow-up samples, were predicted in the OPLS-DA model. 70% of the samples were predicted as Parkinson’s disease (PD), and 23 of 40 individuals had all their longitudinal samples predicted as PD. In the more refined support vector machine (SVM) model, 79% of the 146 new samples were predicted as PD and 27 of 40 individuals consistently had all their longitudinal samples predicted as PD. Source data are provided as a Source Data file.
The correlation between differentially expressed protein biomarkers and patients’ clinical data in the targeted proteomic validation phase (phase I)
We next evaluated the relationship between proteins and clinical data by correlating the protein expression in PD and HC (from phase I) with clinical scores (Mini-Mental State Examination [MMSE], Hoehn & Yahr stage [H&Y] and UPDRS [Unified Parkinson’s Disease Rating Scale; I–III, and total score]). We found negative correlations for GRN, DKK3, PPP3CB, and SELE with H&Y and UPDRS parts II, III, and total score, possibly indicating a connection between a more severe clinical (especially motor) impairment and lower expression of markers in the Wnt-signalling pathways (DKK3 and PPP3CB). Higher Cystatin C plasma levels correlated with higher numbers in UPDRS part III (motor performance) and UPDRS total score. The same was found for PTGDS plasma levels, which were also negatively correlated with MMSE. The central complement cascade protein, C3, negatively correlated with MMSE, and positively correlated with H&Y, UPDRS part III, and total score. The UPR-regulating protein BiP (HSPA5) correlated negatively with MMSE, and positively with H&Y and UPDRS parts II, III, and total score. The ERAD-associated proteins, HSPAIL and adiponectin, were positively correlated with H&Y, and UPDRS parts II, III, and total score. SERPINs (SERPINA3, SERPINF2 and SERPING1) and hemopexin (HPX) correlated negatively with MMSE and positively with H&Y and UPDRS parts II, III, and total score. In general, the MMSE score was inversely correlated with H&Y stage and UPDRS scores. For detailed information, see Fig. 8 and Table 2 .

The correlation was performed using Spearman’s procedure, and the clustering method was set to average. The clustering metric was Euclidean. The heatmap is coloured by correlation coefficient where red represents positive and blue negative correlations. The proteins are represented by gene names. Detailed information about the protein correlations can be found in Supplementary Table 3 . De novo Parkinson’s disease ( n = 99) and healthy controls ( n = 36). MMSE mini-mental state examination, UPDRS unified Parkinson’s disease rating Scale. Source data are provided as a Source Data file.
Comparison of clinical outcomes and measurements in the longitudinal iRBD cohort-Independent replication cohort-phase II
The longitudinal expression in the iRBD samples was evaluated using linear mixed-effects models. Conditional growth models with random slopes and random intercepts between the individuals were constructed. After adjusting the p values for multiple testing by applying the Benjamini–Hochberg (BH) procedure with alpha = 0.05, we found that Butyrylcholinesterase (BCHE) was significantly decreased over the timepoints in the iRBD individuals ( p = 0.01). We next focused only on the iRBD samples with at least two timepoints and for which PD had consistently been predicted in the SVM model ( n = 90). This produced comparable results to the initial model with BCHE significantly related with time since baseline ( p = 0.01), but also TUBA4A was nominally significantly increased ( p = 0.04) although not passing the BH FDR threshold. The modelling also demonstrated that the clinical measurements H&Y ( p = 0.02), UPDRS I–III ( p = 0.02), and UPDRS I and III ( p = 0.03 and 0.03, respectively), were significantly related to the time since baseline in the iRBD group post multiple testing correction. PD non-motor symptoms, as measured on the PD NMS sum score, were strongly correlated with longitudinal motor progression ( p = 5E −8 ). Similarly, the questionnaire for quality of life PDQ-39’s mean values also correlated with longitudinal motor progression ( p = 0.005). From available routine blood values, cholesterol was associated with longitudinal timepoints ( p = 0.02). Details can be found in Supplementary Table 4 . Correlating the clinical measurements with the targeted proteomic data, we applied Spearman’s correlation and found that cholesterol was positively correlated with six of the identified proteins (Supplementary Table 5 ), including HSPA8, APOE and MASP2 ( p = 5E −9 , 0.0003 and 0.003, respectively). Also significantly correlated, but to a lesser degree and not passing the BH FDR threshold, were the PD NMS sum which correlated negatively with TUBA4A (p unadjusted = 0.01) and the PDQ-39 mean values, which correlated negatively with CST3 and PTGDS ( p unadjusted = 0.03 and 0.05, respectively).
PD has emerged as the world’s fastest-growing neurodegenerative disorder and currently affects close to 10 million people worldwide. Consequently, there is an urgent need for disease-modifying and prevention strategies 22 , 23 . The development of such strategies is hampered by two limitations: there are major gaps in our understanding of the earliest events in the molecular pathophysiology of PD, and we lack reliable and objective biomarkers and tests in easily accessible bio-fluids. We, therefore, need biomarkers that can identify PD earlier, preferably a significant time before an individual develops significant neuronal loss and disabling motor and/or cognitive disease. Such biomarkers would advance population-based screenings to identify individuals at risk and who could be included in upcoming prevention trials.
In the last years, CSF SAA emerged as the most specific indicator for NSD, in prodromal stages like iRBD, with an impressively high sensitivity and specificity of up to 74 and 93%, respectively, across various cohorts 9 , 24 . Despite the many questions surrounding SAA that need to be answered, including the ultimate understanding of its functionality, it is a true milestone for advancing prevention trials. It is, however, hampered by having only been shown to be robust in CSF and by the slow development and high variability of SAA in peripheral blood 25 , as well as by the lack of quantification capabilities. An easier and more accessible biofluid test would enable screening large population-based cohorts for at-risk status to develop an NSD. Therefore, the identification of additional biomarkers is needed, as is further knowledge of the biomarkers and pathways of the underlying pathophysiology (e.g. inflammation) during the earliest stage of NSD.
Other emerging multiplex technologies are increasingly used to identify individual proteomic biomarkers. However, these techniques are not true proteomic or ‘eyes open’ methods, as they rely on selected large panels of specific antibodies/and other (e.g. aptamer)-based assay technologies. These techniques, although useful, have not provided consistent results 3 , 26 . Proteomics using mass spectrometry measures all expressed proteins in an unbiased fashion as opposed to those selectively included in a panel that also includes variability due to cross-reactivity. Therefore, proteomic screening using mass spectrometry-based techniques is much more likely to identify pathways or biomarkers and provides more meaningful insights into the disease mechanisms involved in PD. We found a discrepancy between the detected markers during the discovery and the targeted phases. This is a known phenomenon in biomarker translation 27 that is also reflected in the low number of biomarkers having received FDA approval 28 . We addressed this by using previously reported successful improvement strategies in proteomic approaches, namely by refining our panel, reducing the number of markers, and increasing the sample size 29 . Furthermore, the validation of potential biomarkers was performed on a second and different type of mass spectrometer (triple quadrupole), which has the advantage of being available in all large hospitals.
Targeted MS has been previously applied in PD, including by the current authors, but the biological fluid used in the majority of studies is CSF 30 and not peripheral fluids such as blood. Here we demonstrate that even with a very low required volume of plasma/serum (10 µl) targeted proteomic is feasible.
The targeted proteomic assay presented here was developed from proteins identified in an unbiased discovery study, from our previous research, and from the literature. It included several inflammatory markers, Wnt-signalling members, and proteins indicative of protein misfolding. When analysing PD, OND, iRBD and HC in the targeted proteomic validation phase, we identified and confirmed 23 distinct and differentially expressed proteins between PD and HC. Our analysis moreover demonstrated that iRBD possesses a significantly different protein profile compared to HC, consisting of decreased levels of GRN and MASP2 and increased levels of the complement factor C3 and SERPINs (SERPINA3, SERPINF2 and SERPING1), thus indicating early involvement of inflammatory pathways in the initial pathophysiological steps of PD. Comparing these results to previous findings by our and other groups 8 , 31 highlights the link between these proteins and the pathways of complement activation, coagulation cascades, and Wnt-signalling.
By applying machine-learning models, we classified and separated de novo PD or control samples with 100% accuracy based on the expression of eight proteins (GRN, MASP2, HSPA5, PTGDS, ICAM1, C3, DKK3 and SERPING1).
With an independent validation, we added (a) a larger sample set and (b) longitudinal samples from the most interesting subgroup with 54 iRBD subjects and a total of 146 serum samples. We were able to validate our previous panel with a high prediction rate (79%) of these individuals as seen in PD in the targeted approach. Interestingly, the biomarker panel itself did not correlate with longitudinal expression but remained robust after the initial classification of iRBD. So far, 16 of the 54 iRBD subjects converted to PD/DLB (stage 3 NSD). Out of these samples, the SVM model predicted ten individuals with all their timepoints classified as PD, and of the 11 iRBD subjects who converted to PD/DLB, eight were identified as PD by the proteome analysis. Our panel, therefore, identified a PD-specific change in blood up to 7 years before the development of the stage 3 NSD.
The main shortcoming with many previously explored PD biomarkers is weak or no correlation with clinical progression data. So far, outcome measures in clinical trials are primarily based on motor progression, often by a clinical rating scale such as the UPDRS and/or wearable technologies. More objective biomarkers correlating with or reflecting the progression of the pathophysiology and clinical symptoms would be of the utmost importance. We, therefore, calculated correlations with clinical parameters and identified an association with multiple markers, including DKK3, PPP3CB and C3, indicating downregulation of Wnt-signalling pathways. Increased activity of the complement cascade correlated with higher scores in symptom severity (UPDRS part III and total score) and lower scores in cognitive performance (MMSE).
Protein (i.e. α-synuclein) misfolding is a well-known component of PD pathology and is believed to be the key factor behind Lewy body formation 32 . The transport of excessive amounts of misfolded proteins or increased folding cycles can induce ER stress. A cellular defence mechanism to alleviate ER stress is the unfolded protein response (UPR) reducing ER protein influx and increasing protein folding capacity 33 . The UPR is mainly activated by BiP-bound misfolded proteins 34 . The higher expressed markers HSPA5 (UPR-regulating protein BiP) and HSPA1L in our plasma samples of early PD indicate ER stress as a significant factor in the disease process and has been previously linked to PD in both mouse models and brain tissue studies 35 , 36 .
As mentioned by other groups and confirmed in our results, increasing evidence suggests inflammation is a specific feature in early PD. Complement activation has been associated with the formation of α-synuclein and Lewy bodies in PD and deposits of the complement factors iC3b and C9 have been found in Lewy bodies 37 . C3 is a central molecule in the complement cascade and was highly upregulated in blood in both PD and both independent iRBD sample sets analysed in this study. This upregulation in the earliest phase of motor PD (stage 3 NSD), and even in the prodromal phase (stage 2 NSD), clearly indicates inflammation as an early, if not the initial, event in PD neurodegeneration. Complement C3 levels correlated positively with indicators of motor dysfunction (H&Y stage and UPDRS)—indicating a direct connection between high plasma levels of inflammatory proteins and motor symptoms—and negatively with cognitive decline, here with the MMSE.
The protein Mannan-binding serine peptidase 2 (MASP2), an initiator of the lectin part of the complement cascade, was significantly downregulated in PD and iRBD. MASP1 and MASP2 proteins are inhibited by plasma protease C1 inhibitor SERPING1 in the lectin pathway, with SERPING1 modulating the complement cascade as it belongs to the SERPIN family of acute phase proteins 38 . In experimental PD mice models, increased SERPING1 levels are associated with dopaminergic cell death 39 . Acting as a serine/cysteine proteinase inhibitor, SERPING1 can increase serine levels, which could also affect αSyn phosphorylation. This can play a crucial role in PD pathology, as almost 90% of αSyn in Lewy bodies is phosphorylated on Serine129 40 , 41 . We identified increased SERPING1 plasma levels in both PD and iRBD in our analysis (compared to HC), thus contributing to conditions with increased αSyn phosphorylation, consecutive aggregation, Lewy body formation, and finally degeneration of dopaminergic neurons. Furthermore, we observed a strong correlation of SERPING1 plasma levels with UPDRS II, III and total score, as a direct measure of dopaminergic cell loss 39 .
Alpha-2-antiplasmin (SERPINF2) was also significantly upregulated in PD and iRBD. SERPINF2 is a major regulator of the clotting pathway, acting as an inhibitor of plasmin, a serine protease formed upon the proteolytic cleavage of its precursor, plasminogen, by tissue-type plasminogen activator (t-PA) or by the urokinase-type plasminogen activator (u-PA). Plasmin has been reported to cleave and degrade extracellular and aggregated αSyn 42 . Recently, we showed that activation of the plasminogen/plasmin system is decreased in PD, indicated by decreased plasma levels of uPA and its corresponding receptor uPAR, while t-PA was associated with faster disease progression 8 . The upregulation of SERPINF2 observed here is another indicator of decreased plasmin activity. Alpha-1-antichymotrypsin (SERPINA3), a third member of the SERPIN family, was also upregulated in the PD subjects. In the CNS, the primary source of SERPINA3 is astrocytes, where its expression is upregulated by various inflammatory receptor complexes 38 .
Overall, independent upregulation of these three members of the SERPIN (SERPING1, SERPINF2, SERPINA3) family is also indicative of increased inflammatory activity, combined with less activation of the plasmin system, and correlation with motor and non-motor symptom severity. In addition, a strong downregulation of progranulin ( GRN ) was detected, indicating a potential loss of neuroprotection and increased susceptibility to neuroinflammation. GRN may act as a neurotrophic factor, promoting neuronal survival and modulating lysosomal function. Loss-of-function mutations in the GRN gene are a cause of frontotemporal dementia and familial DLB. GRN gene variants are also known to increase the risk of developing Alzheimer’s disease (AD) and PD 43 . The main characteristics of neurodegeneration related to GRN are TDP43(-Transactive response DNA binding protein 43) inclusions, but Lewy body pathology is also very common. Loss of progranulin has further been linked to increased production of pro-inflammatory species such as tumour necrosis factor (TNF) and IL-6 in microglia 15 . A study in mice showed that Grn -/- mice had elevated levels of complement proteins, including C3, even before the onset of neurodegeneration 44 . Additionally, previous studies have found GRN downregulated in serum samples of advanced PD compared to AD and healthy individuals 45 .
As a possible compensatory reaction to the described increased inflammatory markers, the levels of Prostaglandin-H 2 d -isomerase (PTGDS)/Prostaglandin-D 2 synthase (PGDS2), better known as β-trace protein, were upregulated. PDGDS is an important brain enzyme producing prostaglandin D2 (PGD2), which has a neuroprotective and anti-inflammatory function. The upregulation reported here could be a reaction to the amount of neuronal cell loss, which is also seen in the significant correlation with the clinical motor and cognitive scales (see below). Furthermore, β-trace protein is a marker for CSF and is used to identify the fluid in clinical routine diagnostics, thus helping detect CSF leakage 46 . Increased plasma levels could be indicative of a disrupted blood–brain barrier (BBB), often discussed in PD pathology 47 and demonstrated in our cohorts.
Our study shows that the Wnt-related proteins DKK3 and PPP3CB are strongly downregulated in de novo PD. DKK3 is an activator of the canonical Wnt/β-catenin branch and PPP3CB is a component of the non-canonical Wnt/Ca 2+ signalling pathway. Wnts are secreted, cysteine-rich glycoproteins that act as ligands to locally stimulate receptor-mediated signal transduction of the Wnt-pathway 48 . Wnt-signalling is crucial for the development and maintenance of dopaminergic neurons 49 , shows protective effects on midbrain dopaminergic neurons 50 , and seems to be involved in the maintenance of the BBB 48 , 51 . Wnt-ligands and agonists trigger a “Wnt-On” stage, characterised by neuronal plasticity and protection, while the opposite “Wnt-Off” stage, potentially leading to neurodegeneration, triggered by the phosphorylation activity of glycogen synthetase kinase-3β (GSK-3beta) 50 , 52 . Wnt-inhibitors are separated into secreted Frizzled-related proteins (sFRP) and Dickkopf proteins (DKK). DKK1, DKK2 and DKK4 act as antagonists, while DKK3 is an agonist and activator 53 . Adult neurogenesis is primarily governed by canonical Wnt/β-catenin signaling 54 and downregulation of Wnt-signalling promotes dysfunction and/or death of dopaminergic neurons. Restoration of dopaminergic neurons was shown in mice where β-catenin was activated in situ 52 and neural stem cells transplanted to the substantia nigra of medically PD-induced mice induced re-expression of Wnt1 and repair dopaminergic neurons 55 . DKK3 and PPP3CB were strongly downregulated in de novo PD, removing an important line of defence against the detrimental loss of dopaminergic neurons. The downregulation of the Wnt-signalling pathways was further correlated with higher motor scores (UDPRS and H&Y stages).
Wnt-signalling in PD is not only promising as a potential biomarker. In oncology, drugs can modify Wnt-pathways, which is of interest to the PD field 56 . Some substances show no BBB-permeability. As a disrupted BBB seems to be apparent in PD, these drugs may be effective. Furthermore, these substances are also relevant for PD treatment: research points towards a peripheral starting point of PD and future therapies should be administered as early as possible 57 . These promising substances include DKK- as well as GSK inhibitors, but to date, no drugs targeting the Wnt-signalling pathways have been effectively tested in clinical trials, including in those with neurodegenerative diseases. Progress and clinical trials are urgently needed here.
The transfer of multi-omics analysis to clinically meaningful results that directly impact future drug trial planning and biomarker validation, depends fundamentally on correlating these results and altered pathway regulations with established clinical scores. The markers we analysed in our targeted mass spectrometry panel did not only show different expression patterns between HC, PD, and in both of our independent iRBD sample sets, but most of the markers also robustly correlated with important clinical scores (UPDRS and MMSE, see Table 1 ). Cognitive decline correlated negatively with the SERPINs and complement factor C3. The burden of motor and non-motor symptoms and overall symptom severity rated by UPDRS and its subscores correlated positively with the SERPINs, Complement C3, and negatively with DKK3, GRN, and SELE. So, increased inflammatory activity and downregulation of Wnt-signalling seem to strongly affect the clinical picture of PD subjects.
The iRBD subjects showed decreased levels of BCHE over time compared to controls. BCHE has been reported as decreased in serum samples of PD with cognitive impairment 58 . Validation of this easily assessable marker in serum is needed to evaluate its predictive potential.
While we did not find significant differences when we compared paired serum and plasma samples; the analysis of paired samples of plasma/serum and CSF only correlated weakly with the marker concentrations in these peripheral and central compartments. This discrepancy has been reported by several groups 20 , 21 . One reason is that mass spectrometry-based proteome analysis is always biased towards quantification and detection of the most abundant proteins in each sample matrix, and the total protein concentrations in human plasma/serum are more than two orders of magnitude higher than that in CSF. Further, the regulatory function of the blood–brain barrier seems to play a different role for different proteins, as some, like c-reactive protein, show a strong correlation between CSF and plasma, but most of the proteins do not. CSF and blood proteome show complex dynamics influenced by multiple and still mostly unknown factors. The protein shift in samples with a known BBB dysfunction (determined by the CSF/serum albumin index or the CSF/plasma ratio) can not be determined for individual proteins nor the dysfunction be localised by mass spectrometry 20 .
Our model could not correctly predict phenoconversion in all cases. The reasons for this can be varied: The proteome pattern changes over time and the period between sampling and phenconversion may play a role. The three PD phenoconverters that were not predicted as PD neither differ clinically or demographically from the phenoconverters, nor from the non-phenoconverters. iRBD diagnosis in our study was confirmed by vPSG, supported by a high percentage of additional measurements including hyposmia and CSF SAA positivity. Therefore, even those iRBD cases that do not show the PD-proteome pattern still have a high-risk constellation of converting to PD/DLB on three different levels (PSG, olfaction, and SAA). Continuing further longitudinal follow-up of these subjects will elucidate our understanding of when and potentially why conversion occurs/does not occur. It is known that around 80% of iRBD subjects develop NSD, i.e. PD/DLB, with a rate of 6% per year, as shown in a multicenter cohort including ours 59 . To a lesser extent, iRBD subjects develop the intracytoplasmic glial α-synuclein aggregation disorder Multiple Systems Atrophy (MSA) 59 , 60 . Although RBD is common in MSA (summary prevalence of 73% 61 ), none of our iRBD subjects have, as yet converted to MSA. Recruiting and following large longitudinal at-risk cohorts is, therefore, very important and future studies will not only identify biomarkers for phenoconversion from stage 1 or 2 to eventually stage 3 NSD or MSA, but also identify the many possible factors of resilience (including genetics, etc.) of NON-conversion which will be as, if not more important than identifying indicators for phenoconversion. Both direction progression biomarkers from stage 1 and 2 cohorts will have tremendous implications for future neuroprevention trials as phenoconversion itself is (due to the low annual rate) unlikely to be an outcome measure.
A significant strength of our biomarker discovery to translation pipeline is that it allows for the developed test to be easily validated and translated to any clinical laboratory equipped with a tandem LC-MS instrument. One advantage of using triple quadrupole platforms is that additional and better biomarkers can easily be augmented into the test described in this manuscript. Thus, any test could be refined and optimised over time with very little modification to the assay as additional biomarkers are discovered. Clinical testing for neurological disorders is limited to the use of a selected few well-characterised individual markers and translating biomarkers to eventual clinical application is notoriously challenging. The power of using multiplexed biomarker technologies with machine learning enables biomarkers to be evaluated in context with other markers of pathological events, thereby creating a ‘disease profile’ as opposed to individual markers. This approach opens the biomarker discovery field for many disorders and increases the specificity and sensitivity of testing, as demonstrated in this study. The combination of multiplexed analysis of biomarker panels analysed on triple quadrupole platforms can advance biomarker translation to clinical application; this mass spectral technology is already embedded in many clinical diagnostics labs for routine small molecule analyses.
Our peripheral blood protein pattern for PD helps not only to classify but also to predict the earliest stage of the disease. We find differently expressed proteins in pre-motor iRBD and early motor stages of the disease compared to HC. Multiple markers also correlated with the progression of motor and non-motors symptoms. Thus, our blood panel can also identify subjects at risk (stage 2) to develop PD up to 7 years before advancing to motor stage 3. Next steps will be the independent validation in other (and even earlier) non-motor cohorts, e.g. in subjects with hyposmia also at-risk for PD 62 and in our population-based Healthy Brain Ageing cohort in Kassel 63 . It would further be interesting to evaluate the predictive potential of these identified markers with continuing clinical follow-up and together with other established PD progression markers like serum neurofilament light chain 5 and dopamine transporter imaging in a longitudinal analysis.
Our work was predominantly focused on the similarities between PD and iRBD. The authors are unaware of any study that has analysed longitudinally collected samples and prodromal cohorts, including iRBD and phenoconverters. Future work would include (i) validation of our findings in independent cohorts consisting of iRBD and other at-risk subjects for the synuclein aggregation disorders in neurons (PD, DLB) and oligodendrocytes (MSA), (ii) refinement of the panels of biomarkers developed in this study including sensitivity and technical performance, (iii) and using the pipeline described in this manuscript, the identification and validation of additional biomarkers that could distinguish between the different clinical syndromes with the ultimate goal of identifying progression biomarkers as outcome measures for prevention trials.
In summary, instead of single biomarkers, in a univariate approach, we have created a pipeline using a targeted proteomic test of a multiplexed panel of proteins, together with machine learning. This powerful combination of multiple well-selected biomarkers with state-of-the-art machine-learning bioinformatics, allowed us to use a panel of eight biomarkers that could distinguish early PD from HC. This biomarker panel provided a distinct signature of protective and detrimental mechanisms, finally triggering oxidative stress and neuroinflammation, leading to α-synuclein aggregation and LB formation. Moreover, this signature was already present in the prodromal non-motor (stage 2 NSD), up to 7 years before the development of motor/cognitive symptoms (stage 3), supporting the high specificity of iRBD and its high conversion rate to PD/DLB 18 . Most importantly, this blood panel can, in the future, upon further validation help identify subjects at risk of developing PD/DLB and stratify them for upcoming prevention trials.
Patient cohorts and sample collection and processing
Our research complies with all relevant ethical regulations. Institutional review board statements were obtained from the University Medical Centre in Goettingen, Germany, Approval No. 9/7/04 and 36/7/02. The study was conducted according to the Declaration of Helsinki, and all participants gave written informed consent. All plasma, serum and CSF samples from subjects were selected from known cohorts using identical sample processing protocols designed by the Movement Disorder Center Paracelsus-Elena-Clinic.
Patients with de novo PD were diagnosed according to the UK Brain Bank Criteria, without PD-specific medication. Diagnosis in all subjects was supported by (1) a positive (i.e. >30% improvement of UPDRS III after 250 mg of levodopa) acute levodopa challenge testing 64 in all PD subjects, (2) hyposmia by smell identification test (Sniffin Sticks 65 ) in all PD subjects and (3) 1.5-tesla Magnetic Resonance Imaging (MRI) without significant abnormalities or evidence for other diseases in all but three subjects who were excluded (due to significant vascular lesions or evidence for hydrocephalus) from the analysis. Participants not fulfilling the above criteria and meeting criteria for other neurological disorders were named as other neurological disorders (OND). OND consists of subjects with vascular parkinsonism ( n = 10), essential tremor ( n = 7), progressive supranuclear palsy; PSP ( n = 7), multiple system atrophy; MSA ( n = 3), corticobasal syndrome; CBS ( n = 2), DLB ( n = 2), drug-induced tremor ( n = 2), dystonic tremor ( n = 2), restless legs syndrome ( n = 1), hemifacial spasm ( n = 1), motoneuron disease ( n = 1), amyotrophic shoulder neuralgia ( n = 1), and Alzheimer’s disease ( n = 1). The initial exploratory cohort consisted of ten PD subjects (8 men, mean age 67.1 ± 10.6) and ten healthy controls (5 men, mean age 65,7, SD ± 8,6.). For details, see Supplementary Table 3 ). The validation cohort included 99 PD subjects (49 men, mean age 66,1, SD ± 10,8), 36 healthy controls (20 men, mean age 63.7, SD ± 6,5.) and the described (see above) 41 OND subjects (29 men, mean age 70, SD ± 8.9. For details, see Supplementary Table 1 . The prodromal validation cohort consisted of 54 patients with iRBD (27 men, mean age 67.5, SD ± 8.1, for details, see Supplementary Table 4 ). RBD was diagnosed with two nights of state-of-the-art vPSG. Samples from HC were selected from the DeNoPa cohort 10 and matched for age and sex with the PD patients, had to be between 40 and 85 years old, without any active known/treated CNS condition, and with a negative family history of idiopathic PD. Antipsychotic drugs were an exclusion criterion. The provided data for sex are based on self-report.
The paired sample analysis of CSF, plasma and serum was applied in samples from subjects with OND 7 men, mean age 74 years, SD ± 7; diagnosis: four Alzheimer’s disease, three vascular Parkinsonism, one essential tremor, one multiple system atrophy one progressive supranuclear palsy).
Clinical assessments included the UPDRS subscores (parts I–III), the sum (UPDRS total score), and cognitive screening using the MMSE 10 .
Plasma and serum samples for both cohorts were collected in the morning under fasting conditions using Monovette tubes (Sarstedt, Nümbrecht, Germany) for EDTA plasma and serum collection by venipuncture. Tubes were centrifuged at 2500× g at room temperature (20 °C) for 10 min and aliquoted and frozen within 30 min of collection at −80 °C until analysis 10 , 66 . Single- use aliquots were used for all analyses presented here. For further details, we refer to the following publication 67 .
CSF was collected in polypropylene tubes (Sarstedt, Nümbrecht, Germany) directly after the plasma collection by lumbar puncture in the sitting position. Tubes were centrifuged at 2500× g at room temperature (20 °C) for 10 min and aliquoted and frozen within 30 min after collection at −80 °C until analysis. Before centrifugation, white and red blood cell counts in CSF were determined manually 10 , 66 . CSF β-amyloid 1–42, total tau protein (t-tau), phosphorylated tau protein (p-tau181) and neurofilament light chains (NFL) concentrations were measured by board-certified laboratory technicians, who were blinded to clinical data, using commercially available INNOTEST ELISA kits for the tau and Aβ markers (Fujirebio Europe, Ghent, Belgium) and the UmanDiagnostics NF-light® assay (UmanDiagnostics, Umeå, Sweden) for NFL. Total protein and albumin levels were measured by nephelometry (Dade Behring/Siemens Healthcare Diagnostics) 66 .
For the α-synuclein seeding aggregation assay (αSyn-SAA) the CSF samples were blindly analyzed in triplicate (40 μL/well) in a reaction mixture (0.3 mg/mL recombinant α-Syn (Amprion [California, USA]; catalogue number S2020), 100 mM piperazine- N , N ′-bis(2-ethanesulfonic acid) (PIPES) pH 6.50, 500 mM sodium chloride, 10 μM thioflavin T, and one bovine serum albumin (BSA)–blocked 2.4-mm silicon nitride G3 bead (Tsubaki-Nakashima [Georgia, USA]). Beads were blocked in 1% BSA 100 mM PIPES pH 6.50 and washed with 100 mM PIPES pH 6.50. The assay was performed in 96-well plates (Costar [New York, USA], catalogue number 3916) using a FLUOstar Omega fluorometer (BMG [Ortenberg, Germany]). Plates were orbitally shaken (800 rpm for 1 min every 29 min at 37 °C). Results from the triplicates were considered input for a three-output probabilistic algorithm with sample labelling as “positive,” “negative,” or “inconclusive”, based on the parameters: Maximum fluorescence (Fmax), time to reach 50% Fmax (T50), slope, and the coefficient of determination for the fitting were calculated for each replicate using a sigmoidal equation available in Mars data analysis software (BMG). The time to reach the 5000 relative fluorescence units (RFU) threshold (TTT) was calculated with a user-defined equation in Mars 19 .
Discovery plasma proteomics (phase 0)
In the mass spectrometry-based proteomic discovery analysis of plasma, we depleted the control and de novo PD samples from the twelve most abundant plasma proteins using Pierce Top12 columns (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The depleted samples were freeze-dried before the addition of 20 µL of lysis buffer (100 mM Tris pH 7.8, 6 M urea, 2 M thiourea, and 2% ASB-14). The samples were shaken on an orbital shaker for 60 min at 1500 rpm. To break disulphide bonds, 45 µg DTE was added, and the samples were incubated for 60 min. To prevent disulphide bonds from reforming, 108 µg IAA was added, and the samples were incubated for 45 min covered in light. About 165 µL MilliQ water was added to dilute the concentration of urea and 1 µg trypsin gold (Promega, Mannheim, Germany) was added before 16 h of incubation at +37 °C to digest the proteins into peptides. To purify the peptides, solid phase extraction was performed using 100 mg C18 cartridges (Biotage, Uppsala, Sweden). The cartridges were washed with two 1 mL aliquots of 60% ACN, and 0.1% TFA before equilibration by two 1 mL aliquots of 0.1% TFA. The concentration of TFA in the samples was adjusted to 0.1%. The samples were loaded, and the flow-through was captured and re-applied. Salts were washed away from the bound peptides by two 1 mL aliquots of 0.1% TFA. The peptides were eluted by two 250 µL aliquots of 60% ACN, and 0.1% TFA. Solvents were evaporated using a vacuum concentrator. The samples were re-suspended in 50 µL 3% ACN, 0.1% FA prior to analysis. About 4 µL was injected into a 2D-NanoAquity liquid chromatography system (Waters, Manchester, UK). All samples were fractionated online into ten fractions over 12 h. The mobile phase in the first chromatographic system consisted of A1: 10 mM ammonium hydroxide titrated to pH 9 and B1: acetonitrile. The second chromatographic system’s mobile phase was A2: 5% dimethylsulfoxide (DMSO) + 0.1% formic acid, B2: acetonitrile with 5% DMSO + 0.1% formic acid. 2D-liquid chromatography fractionation was performed by loading the sample onto a 300 µm × 50 mm, 5 µm Peptide BEH C18 column (Waters). The peptides were eluted from the first column at a flow rate of 2 µL/min. The initial condition of the gradient elution was 3% B, held over 0.5 minutes before linearly increasing the proportion of organic solvent B, fraction per fraction over 0.5 min. The conditions thereafter remained static for 4 min before returning to the initial conditions over 0.5 min and equilibration prior to the next elution for 10 min. The eluted peptides from the first-dimensional column were loaded into a 180 µm × 20 mm, 5 µm Symmetry C18 trap column (Waters) before entering the analytical column, a 75 µm × 150 mm, 1.7 µm Peptide BEH C18 (Waters). The column temperature was +45 °C. The gradient elution applied to the analytical column started at 3% B and was linearly increased to 40% B over 40 min after which it was increased to 85% B over 2 min and washed for 2 min before returning to initial conditions over 2 min followed by equilibration for 15 min before the subsequent injection. The eluted peptides were detected using a Synapt-G2-S i (Waters) equipped with a nano-electrospray ion source. Data were acquired in positive MS E mode from 0 to 60 min within the m/z range 50−2000. The capillary voltage was set to 3 kV and the source temperature to +100 °C. The desolvation gas consisted of nitrogen with a flow rate of 50 L/h, and the desolvation temperature was set to +200 °C. The purge and desolvation gas consisted of nitrogen, operated at a flow rate of 600 mL/h and 600 L/h, respectively. The gas in the IMS cell was helium, with a flow rate of 90 mL/h. The low energy acquisition was performed by applying a constant collision energy of 4 V with a 1-s scan time. High energy acquisition was performed by applying a collision energy ramp, from 15 to 40 V, and the scan time was 1 s. The lock mass consisted of 500 fmol/µL [glu1]-fibrinopeptide B, continuously infused at a flow rate of 0.3 µL/min and acquired every 30 s. The doubly charged precursor ion, m/z 785.8426, was utilised for mass correction. After acquisition, data were imported to Progenesis QI for proteomics (Waters), and the individual fractions were processed before all results were merged into one experiment. The Ion Accounting workflow was utilised, with UniProt Canonical Human Proteome as a database (build 2016). The digestion enzyme was set as trypsin. Carbamidomethyl on cysteines was set as a fixed modification; deamidation of glutamine and asparagine, and oxidation of tryptophan and pyrrolidone carboxylic acid on the N-terminus were set as variable modifications. The identification tolerance was restricted to at least two fragments per peptide, three fragments per protein, and one peptide per protein. A FDR of 4% or less was accepted. The resulting identifications and intensities were exported and variables with a confidence score less than 15 and only one unique peptide were filtered out.
Targeted plasma proteomics (phase I)
The peptides included in the targeted assay were selected from several proteomic screening studies in which we analysed plasma, serum, urine, and CSF in ageing, PD and AD. The analytical method is described by ref. 17 . Furthermore, due to the suggested involvement of inflammation in neurodegenerative diseases, several known pro- and anti-inflammatory proteins identified from the literature were included in the multiplexed assay. The final panel consisted of 121 proteins (Supplementary Table 2 ), out of which a number were measured with two peptides, leading to a total of 167 unique peptides. When possible, the peptides were chosen to have an amino acid sequence length between 7 and 20. The amino acid sequences were confirmed to be unique to the proteins by using the Basic Local Alignment Search Tool (BLAST) provided by UniProt 68 . Synthetic peptide standards were purchased from GenScript (Amsterdam, Netherlands). To establish the most optimal transitions, repeated injections of 1 pmol peptide standard onto a Waters Acquity ultra-performance liquid chromatography (UPLC) system coupled to a Waters Xevo-TQ-S triple quadrupole MS were performed. The most high-abundant precursor-to-product ion transitions and their optimal collision energies were determined manually or using Skyline 69 . Detection was performed in positive ESI mode. The capillary voltage was set to 2.8 kV, the source temperature to 150 °C, the desolvation temperature to 600 °C, and the cone gas and desolvation gas flows to 150 and 1000 L/h, respectively. The collision gas consisted of nitrogen and was set to 0.15 mL/min. The nebuliser operated at 7 bar. Two transitions were chosen, one quantifier for relative concentration determination and one qualifier for identification, totally rendering 334 analyte transitions. Cone and collision energies varied depending on the optimal settings for each peptide. Each peptide was measured with a minimum of 12 points per peak and a dwell time of 10 ms or more to ensure adequate data acquisition. The optimised transitions were distributed over two multiple reaction monitoring (MRM) methods, always keeping the quantifier and qualifier for each peptide in the same MRM segment. Plasma, serum, and CSF samples were depleted from albumin and IgG using Pierce Top2 cartridges (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. About 150 µg whole protein yeast enolase (ENO1) was added to the cartridges as an internal standard to account for digestion efficiency. Digestion was performed as described above. Solid phase extraction was carried out on BondElute 100 mg C18 96-well plates (Agilent, Santa Clara, USA) using the same methodology as in the preparation of untargeted proteomic analyses. Quality control samples were prepared from acetone-precipitated plasma, digested and solid phase extracted. Calibration curves ranging from 0 to 1 pmol/μL were constructed in blank and matrix by spiking increasing amounts of peptides into blank and QC samples. Before analysis, the samples were reconstituted in 30 µL 3% ACN, 0.1% FA containing 0.1 μM heavy isotope labelled peptides from the following proteins (annotated by gene name): ALDOA, C3, GSTO1, RSU1 and TSP1. About 5 µL were injected. The peptides were separated and detected on an Acquity UPLC system coupled to a Xevo-TQ-S triple quadrupole mass spectrometer (Waters, Manchester, UK). Chromatographic separation of the peptides was performed using a 1 × 100 mm, 1.7 μm ACQUITY UPLC Peptide CSH C18 column (Waters).
The mobile phase consisted of A: 0.1% formic acid and B: 0.1% formic acid in acetonitrile pumped at a flow rate of 0.2 mL/min. The column temperature was set to +55 °C. The initial mobile phase composition was 3% B, which was kept static for 0.8 min before initialising the linear gradient, running for 7.6 min to 25% B, eluting most of the peptides. B was thereafter linearly increased to 80% over 0.5 min and held for 1.9 min, eluting the most apolar peptides and washing the column before returning to the initial conditions over 0.1 minutes followed by equilibration for 6 min prior to the subsequent injection. Two subsequent injections of each sample were performed, each paired with one of the two MRM acquisition methods.
After acquisition, peak-picking and integration were performed using TargetLynx (version 4.1, Waters) or an in-house application ('mrmIntegrate') written in Python (version 3.8). mrmIntegrate is publicly available to download via the GitHub repository https://github.com/jchallqvist/mrmIntegrate . The application takes text files as input (.raw files are transformed into text files through the application 'MSConvert' from ProteoWizard 70 and applies a LOWESS filter over five points of the chromatogram. The integration method to produce areas under the curve is trapezoidal integration. The application enables retention time alignment and simultaneous integration of the same transition for all samples. Peptide peaks were identified by the blank and matrix calibration curves. The integrated peak areas were exported to Microsoft Excel, where first, the ratio between quantifier and qualifier peak areas were evaluated to ensure that the correct peaks had been integrated. The digestion efficiency was evaluated by monitoring the presence of baker’s yeast ENO1 in the samples, all samples without a signal were excluded from further analysis. After the initial quality assessment, the quantifier area was divided by the area of one of the internal standards, ALDOA or GSTO1 to yield a ratio used for the determination of relative concentrations. Any compound that also showed an intensity signal in the blank samples had the blank signal subtracted from the analyte peak intensity. Pooled plasma quality control samples were additionally evaluated to assess the robustness of the run.
Refined LC-MS/MS method (phase II)
The rapid and refined targeted proteomics LC-MS/MS method contained only peptides from the 31 proteins observed in the original targeted proteomics method (121 proteins). We utilised a Waters Acquity (UPLC) system coupled to a Waters Xevo-TQ-XS triple quadrupole operating in positive ESI mode. The column was an ACQUITY Premier Peptide BEH C18, 300 Å, 1.7 µm, maintained at 40 °C. The mobile phase was A: 0.1% formic acid in water, and B: 0.1% formic acid in acetonitrile. The gradient elution profile was initiated with 5% B and held for 0.25 min before linearly increasing to 40% B over 9.75 min to elute and separate the peptides. The column was washed for 1.6 min with 85% B before returning to the initial conditions and equilibrating for 0.4 min. The flow rate was 0.6 mL/min. The settings of the mass spectrometer and the peak-picking method were the same as described in the prior section. Baker’s yeast ENO1 was utilised to monitor digestion efficiency and as an internal standard.
Statistical methods
Most of the statistical analyses were performed in Python (version 3.8.5). The untargeted and targeted datasets were inspected for outliers and instrumental drift using principal component analysis (PCA) and orthogonal projection to latent variables (OPLS) in SIMCA, version 17 (Umetrics Sartorius Stedim, Umeå, Sweden). Outliers exceeding ten median deviations from each variable’s median were excluded. Instrumental drift was corrected by applying a non-parametric LOWESS filter from statsmodels (version 0.14.0) using 0.5 fractions of the data to estimate the LOWESS curve 71 . The data were evaluated for normal distribution using D’Agostino and Pearson’s method from SciPy (version 1.9.3) 72 . The non-normally distributed variables in the untargeted data were transformed to normality by the Box-Cox procedure using the SciPy function 'boxcox'. Significance testing between the independent groups of HC and PD/OND/iRBD individuals was performed by Student’s two-tailed t -test for the untargeted proteomic data and by Mann–Whitney’s non-parametric U -test (SciPy) for the targeted data. Due to the limited sample numbers, no multiple testing correction was performed in the untargeted data. In the targeted data, the Benjamini–Hochberg multiple testing correction procedure (statsmodels) was applied with an accepted false discovery rate of 5%. Fold-changes were calculated by dividing the means of the affected groups by the control group. Correlation analyses in the targeted data were performed by Spearman’s correlation (SciPy) and the correlation p values were adjusted variable-wise by the Benjamini–Hochberg procedure (FDR = 5%).
We implemented a support vector classifier model to discriminate between PD and HC and to predict new samples. The data were first z-scored protein-wise and any 'not a number'-values were replaced by the median. We used the 'LinearSVC' method from SciKit Learn and applied cross-validated recursive feature elimination to determine the number of variables to use in the model. The most discriminating variables for distinguishing between controls and PD were thereafter chosen by recursive feature elimination 73 . Feature selection and model training were performed on 70% of the data, partitioned using the SciKit Learn function “train_test_split”, and cross-validation was performed using a stratified k-fold with five splits. The remaining 30% of the data were predicted in the model. PR and ROC curves were constructed from the test data and consisted of each predictor and from the combined predictors, the packages precision_recall_curve and roc_curve from SciKit Learn were implemented. Linear mixed models were performed using the R-to-Python bridge software pymer4 (version 0.8.0), where individual was set as a random effect and the correlations between the MS measured proteins and clinical variables were evaluated for significance post Benjamini–Hochberg’s procedure for multiple testing correction. Plots of the data were constructed using the Seaborn and Matplotlib packages (versions 0.12.2 and 3.6.0, respectively) 74 .
All multivariate analyses were performed in SIMCA, version 17. OPLS and OPLS-discriminant analysis (OPLS-DA) models were evaluated for significance by ANOVA p values and by permutation tests applying 1000 permutations, where p < 0.05 and p < 0.001 were deemed significant, respectively.
Data were analysed for pathway enrichment using IPA (QIAGEN Inc. Data were analysed for pathway enrichment using IPA (QIAGEN Inc., https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ .). Input variables were set to proteins demonstrating a significant difference between PD individuals and HC, with fold-change as expression observation. The accepted output pathways were restricted to p < 0.05 and at least two proteins were included in the pathways. Gene Ontology (GO) annotations were extracted using DAVID Bioinformatics Resources (2021 build) 75 , 76 . Networks were built in Cytoscape 77 (version 3.8.0) by applying the “Organic layout” from yFiles 77 .
Obtaining biological materials
Patient samples can be provided to other researchers for certain projects after contact with the corresponding authors and upon availability approval of the team in Kassel, Germany.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The chromatograms from the targeted mass spectrometric data generated in this study have been deposited in the ProteomeXchange database under accession code PXD041419 and in the Panorama repository ( https://panoramaweb.org/DNP_Pub.url , https://doi.org/10.6069/p9cy-h335 ). The integrated targeted mass spectrometric data generated in this study are provided in the Supplementary Information. Source data for all data presented in graphs within the figures are provided in a source data file. Source data are provided with this paper.
Code availability
Peak-picking and integrations were performed in TargetLynx (part of the MassLynx suite, version 4.1), or using an in-house application written in Python which can be found on GitHub ( https://github.com/jchallqvist/mrmIntegrate ). The data visualisation and statistical analyses were performed in Python (version 3.8.5) using the packages SciPy (version 1.9.3), statsmodels (version 0.14.0), SciKit Learn (version 1.1.2), Seaborn (version 13.0) and Matplotlib (version 3.6.0). The code used can be found on GitHub ( https://github.com/jchallqvist/DNP_Pub/blob/main/DNP_Code , https://doi.org/10.5281/zenodo.11130369 ).
Simuni, T. et al. Baseline prevalence and longitudinal evolution of non-motor symptoms in early Parkinson’s disease: the PPMI cohort. J. Neurol. Neurosurg. Psychiatry 89 , 78–88 (2018).
Article PubMed Google Scholar
Michell, A. W., Lewis, S. J., Foltynie, T. & Barker, R. A. Biomarkers and Parkinson’s disease. Brain 127 , 1693–1705 (2004).
Article CAS PubMed Google Scholar
Kieburtz, K., Katz, R., McGarry, A. & Olanow, C. W. A new approach to the development of disease-modifying therapies for PD; fighting another pandemic. Mov. Disord. 36 , 59–63 (2021).
Shahnawaz, M. et al. Development of a biochemical diagnosis of Parkinson disease by detection of α-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol. 74 , 163–172, (2017).
Mollenhauer, B. et al. Validation of serum neurofilament light chain as a biomarker of Parkinson’s disease progression. Mov. Disord . https://doi.org/10.1002/mds.28206 (2020).
Lindestam Arlehamn, C. S. et al. α-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat. Commun. 11 , 1875 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar
Mollenhauer, B. et al. Baseline predictors for progression 4 years after Parkinson’s disease diagnosis in the De Novo Parkinson Cohort (DeNoPa). Mov. Disord. 34 , 67–77 (2019).
Bartl, M. et al. Blood markers of inflammation, neurodegeneration, and cardiovascular risk in early Parkinson’s disease. Mov. Disord . https://doi.org/10.1002/mds.29257 (2022).
Simuni, T. et al. A biological definition of neuronal α-synuclein disease: towards an integrated staging system for research. Lancet Neurol. 23 , 178–190 (2024).
Mollenhauer, B. et al. Nonmotor and diagnostic findings in subjects with de novo Parkinson disease of the DeNoPa cohort. Neurology 81 , 1226–1234 (2013).
Hällqvist, J. et al. A multiplexed urinary biomarker panel has potential for Alzheimer’s disease diagnosis using targeted proteomics and machine learning. Int. J. Mol. Sci . https://doi.org/10.3390/ijms241813758 (2023).
Hu, W., Ralay Ranaivo, H., Craft, J. M., Van Eldik, L. J. & Watterson, D. M. Validation of the neuroinflammation cycle as a drug discovery target using integrative chemical biology and lead compound development with an Alzheimer’s disease-related mouse model. Curr. Alzheimer Res. 2 , 197–205 (2005).
Notter, T. et al. Translational evaluation of translocator protein as a marker of neuroinflammation in schizophrenia. Mol. Psychiatry 23 , 323–334 (2018).
Jonsson, M., Gerdle, B., Ghafouri, B. & Backryd, E. The inflammatory profile of cerebrospinal fluid, plasma, and saliva from patients with severe neuropathic pain and healthy controls-a pilot study. BMC Neurosci. 22 , 6 (2021).
Article PubMed PubMed Central Google Scholar
Chen, X. et al. Progranulin does not bind tumor necrosis factor (TNF) receptors and is not a direct regulator of TNF-dependent signaling or bioactivity in immune or neuronal cells. J. Neurosci. 33 , 9202–9213 (2013).
Article CAS PubMed PubMed Central Google Scholar
Captur, G. et al. Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection. EBioMedicine 85 , 104293 (2022).
Doykov, I. et al. The long tail of Covid-19’ - The detection of a prolonged inflammatory response after a SARS-CoV-2 infection in asymptomatic and mildly affected patients. F1000Res 9 , 1349 (2020).
Hu, M. T. REM sleep behavior disorder (RBD). Neurobiol. Dis. 143 , 104996 (2020).
Concha-Marambio, L. et al. Accurate detection of α-synuclein seeds in cerebrospinal fluid from isolated rapid eye movement sleep behavior disorder and patients with Parkinson’s disease in the de novo Parkinson (DeNoPa) cohort. Mov. Disord. 38 , 567–578 (2023).
Dayon, L. et al. Proteomes of paired human cerebrospinal fluid and plasma: relation to blood-brain barrier permeability in older adults. J. Proteome Res. 18 , 1162–1174 (2019).
Whelan, C. D. et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. Acta Neuropathol. Commun. 7 , 169 (2019).
Bloem, B. R., Okun, M. S. & Klein, C. Parkinson’s disease. Lancet 397 , 2284–2303 (2021).
Dorsey, E. R., Sherer, T., Okun, M. S. & Bloem, B. R. The emerging evidence of the Parkinson pandemic. J. Parkinsons Dis. 8 , S3–s8 (2018).
Grossauer, A. et al. α-Synuclein seed amplification assays in the diagnosis of synucleinopathies using cerebrospinal fluid-A systematic review and meta-analysis. Mov. Disord. Clin. Pr. 10 , 737–747 (2023).
Article Google Scholar
Okuzumi, A. et al. Propagative α-synuclein seeds as serum biomarkers for synucleinopathies. Nat. Med. 29 , 1448–1455 (2023).
Raffield, L. M. et al. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 20 , e1900278 (2020).
Hernández, B., Parnell, A. & Pennington, S. R. Why have so few proteomic biomarkers “survived” validation? (Sample size and independent validation considerations). Proteomics 14 , 1587–1592 (2014).
Füzéry, A. K., Levin, J., Chan, M. M. & Chan, D. W. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin. Proteom. 10 , 13 (2013).
Bader, J. M., Albrecht, V. & Mann, M. MS-based proteomics of body fluids: the end of the beginning. Mol. Cell Proteom. 22 , 100577 (2023).
Article CAS Google Scholar
Pan, C. et al. Targeted discovery and validation of plasma biomarkers of Parkinson’s disease. J. Proteome Res. 13 , 4535–4545 (2014).
Qin, X. Y., Zhang, S. P., Cao, C., Loh, Y. P. & Cheng, Y. Aberrations in peripheral inflammatory cytokine levels in Parkinson disease: a systematic review and meta-analysis. JAMA Neurol. 73 , 1316–1324, (2016).
Choi, M. L. & Gandhi, S. Crucial role of protein oligomerization in the pathogenesis of Alzheimer’s and Parkinson’s diseases. FEBS J. 285 , 3631–3644 (2018).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 , 1081–1086 (2011).
Article ADS CAS PubMed Google Scholar
Bertolotti, A., Zhang, Y. H., Hendershot, L. M., Harding, H. P. & Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2 , 326–332 (2000).
Colla, E. Linking the endoplasmic reticulum to Parkinson’s disease and alpha-synucleinopathy. Front. Neurosci. 13 , 560 (2019).
Mercado, G., Castillo, V., Soto, P. & Sidhu, A. ER stress and Parkinson’s disease: pathological inputs that converge into the secretory pathway. Brain Res. 1648 , 626–632 (2016).
Loeffler, D. A., Camp, D. M. & Conant, S. B. Complement activation in the Parkinson’s disease substantia nigra: an immunocytochemical study. J. Neuroinflamm 3 , 29 (2006).
Zattoni, M. et al. Serpin signatures in Prion and Alzheimer’s diseases. Mol. Neurobiol. 59 , 3778–3799 (2022).
Seo, M. H. & Yeo, S. Association of increase in Serping1 level with dopaminergic cell reduction in an MPTP-induced Parkinson’s disease mouse model. Brain Res. Bull. 162 , 67–72 (2020).
Anderson, J. P. et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281 , 29739–29752 (2006).
Fujiwara, H. et al. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4 , 160–164 (2002).
Kim, K. S. et al. Proteolytic cleavage of extracellular alpha-synuclein by plasmin implications for Parkinson disease. J. Biol. Chem. 287 , 24862–24872 (2012).
Reho, P. et al. GRN mutations are associated with Lewy body dementia. Mov. Disord. 37 , 1943–1948 (2022).
Kao, A. W., Mckay, A., Singh, P. P., Brunet, A. & Huang, E. J. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18 , 325–333 (2017).
Mateo, I. et al. Reduced serum progranulin level might be associated with Parkinson’s disease risk. Eur. J. Neurol. 20 , 1571–1573 (2013).
Bachmann-Harildstad, G. Diagnostic values of beta-2 transferrin and beta-trace protein as markers for cerebrospinal fluid fistula. Rhinology 46 , 82–85 (2008).
PubMed Google Scholar
Pediaditakis, I. et al. Modeling alpha-synuclein pathology in a human brain-chip to assess blood-brain barrier disruption. Nat. Commun. 12 , 5907 (2021).
Serafino, A., Giovannini, D., Rossi, S. & Cozzolino, M. Targeting the Wnt/β-catenin pathway in neurodegenerative diseases: recent approaches and current challenges. Expert Opin. Drug Discov. 15 , 803–822 (2020).
Arenas, E. Wnt signaling in midbrain dopaminergic neuron development and regenerative medicine for Parkinson’s disease. J. Mol. Cell Biol. 6 , 42–53 (2014).
L’Episcopo, F. et al. A Wnt1 regulated Frizzled-1/β-Catenin signaling pathway as a candidate regulatory circuit controlling mesencephalic dopaminergic neuron-astrocyte crosstalk: therapeutical relevance for neuron survival and neuroprotection. Mol. Neurodegener. 6 , 49 (2011).
Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14 , 133–150 (2018).
L’Episcopo, F. et al. Wnt/beta-catenin signaling is required to rescue midbrain dopaminergic progenitors and promote neurorepair in ageing mouse model of Parkinson’s disease. Stem Cells 32 , 2147–2163 (2014).
Marchetti, B. Wnt/beta-catenin signaling pathway governs a full program for dopaminergic neuron survival, neurorescue and regeneration in the MPTP mouse model of Parkinson’s disease. Int. J. Mol. Sci. 19 , 3743 (2018).
Marchetti, B. et al. Parkinson’s disease, aging and adult neurogenesis: Wnt/beta-catenin signalling as the key to unlock the mystery of endogenous brain repair. Aging Cell 19 , e1310110 (2020).
L’Episcopo, F. et al. Neural stem cell grafts promote astroglia-driven neurorestoration in the aged Parkinsonian brain via Wnt/beta-catenin signaling. Stem Cells 36 , 1179–1197 (2018).
Serafino, A. et al. Developing drugs that target the Wnt pathway: recent approaches in cancer and neurodegenerative diseases. Expert Opin. Drug Discov. 12 , 169–186 (2017).
Harms, A. S., Ferreira, S. A. & Romero-Ramos, M. Periphery and brain, innate and adaptive immunity in Parkinson’s disease. Acta Neuropathol. 141 , 527–545 (2021).
Dong, M. X. et al. Serum butyrylcholinesterase activity: a biomarker for Parkinson’s disease and related dementia. Biomed. Res. Int. 2017 , 1524107 (2017).
Postuma, R. B. et al. Risk and predictors of dementia and parkinsonism in idiopathic REM sleep behaviour disorder: a multicentre study. Brain 142 , 744–759 (2019).
Zhang, H. et al. Risk factors for phenoconversion in rapid eye movement sleep behavior disorder. Ann. Neurol. 91 , 404–416 (2022).
Palma, J. A. et al. Prevalence of REM sleep behavior disorder in multiple system atrophy: a multicenter study and meta-analysis. Clin. Auton. Res. 25 , 69–75 (2015).
Jennings, D. et al. Imaging prodromal Parkinson disease: the Parkinson Associated Risk Syndrome Study. Neurology 83 , 1739–1746 (2014).
Schade, S. et al. Identifying prodromal NMS in a population-based recruitment strategy: Kassel data of Healthy Brain Ageing. Zenodo (2023).
Schade, S. et al. Acute levodopa challenge test in patients with de novo Parkinson’s disease: data from the DeNoPa cohort. Mov. Disord. Clin. Pr. 4 , 755–762 (2017).
Hummel, T., Sekinger, B., Wolf, S. R., Pauli, E. & Kobal, G. ‘Sniffin’ sticks’: olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chem. Senses 22 , 39–52 (1997).
Mollenhauer, B. et al. Monitoring of 30 marker candidates in early Parkinson disease as progression markers. Neurology 87 , 168–177 (2016).
Mollenhauer, B. et al. α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: a cohort study. Lancet Neurol. 10 , 230–240 (2011).
UniProt. BLAST https://www.uniprot.org/blast/ . Accessed January 2024.
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26 , 966–968 (2010).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30 , 918–920 (2012).
Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with Python. In Proc. 9th Python in Science Conference (SciPy, 2010).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17 , 261–272 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12 , 2825–2830 (2011).
MathSciNet Google Scholar
Hunter, J. D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9 , 90–95 (2007).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50 , W216–221, (2022).
Huang Da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 , 44–57 (2009).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 , 2498–2504 (2003).
Download references
Acknowledgements
This work was supported by the Michael J Fox Foundation, PDUK, The Peto Foundation, The TMSRG (UCL), The BRC at Great Ormond Street Hospital, and the Horizon 2020 Framework Programme (Grant number 634821, PROPAG-AGING). We thank the PROPAG-AGING consortium, a full list of the members can be found in the supplementary material.
Open Access funding enabled and organized by Projekt DEAL.
Author information
These authors contributed equally: Jenny Hällqvist, Michael Bartl.
These authors jointly supervised this work: Kevin Mills, Brit Mollenhauer.
Authors and Affiliations
UCL Institute of Child Health and Great Ormond Street Hospital, London, UK
Jenny Hällqvist, Ivan Doykov, Justyna Śpiewak, Héloїse Vinette & Wendy E. Heywood
UCL Queen Square Institute of Neurology, Clinical and Movement Neurosciences, London, UK
Jenny Hällqvist & Kevin Mills
Department of Neurology, University Medical Center Goettingen, Goettingen, Germany
Michael Bartl, Mohammed Dakna, Mary Xylaki, Sandrina Weber & Brit Mollenhauer
Institute for Neuroimmunology and Multiple Sclerosis Research, University Medical Center Goettingen, Goettingen, Germany
Michael Bartl
Paracelsus-Elena-Klinik, Kassel, Germany
Sebastian Schade, Maria-Lucia Muntean, Friederike Sixel-Döring, Claudia Trenkwalder & Brit Mollenhauer
Department of Experimental, Diagnostic, and Specialty Medicine (DIMES), University of Bologna, Bologna, Italy
Paolo Garagnani, Chiara Pirazzini & Claudio Franceschi
IRCCS Istituto delle Scienze Neurologiche di Bologna, Bologna, Italy
Maria-Giulia Bacalini
National Hospital for Neurology & Neurosurgery, Queen Square, WC1N3BG, London, UK
Kailash Bhatia & Sebastian Schreglmann
Institute of Diagnostic and Interventional Neuroradiology, University Medical Center Goettingen, Goettingen, Germany
Marielle Ernst
Department of Neurology, Philipps-University, Marburg, Germany
Friederike Sixel-Döring
UCL: Food, Microbiomes and Health Institute Strategic Programme, Quadram Institute Bioscience, Norwich Research Park, Norwich, UK
Héloїse Vinette
Department of Neurosurgery, University Medical Center Goettingen, Goettingen, Germany
Claudia Trenkwalder
You can also search for this author in PubMed Google Scholar
Contributions
J.H., M.B., K.M., and B.M. conceptualised, planned and oversaw all aspects of the study. J.H., K.M., J.S., H.V., M.B. and S. Schreglmann performed and analyzed most of the experiments. S. Schade, S.W. and M.B. consented to the subjects and collected the samples. M.-L.M., F.S.-D. and S. Schade analyzed the sleep lab data and diagnosed the iRBD subjects. J.H. and M.D. performed the statistical data analysis. J.H. applied the machine learning methods and designed the figures. W.H., I.D., C.F., M.-G.B., P.G., C.P., K.B. and M.X. provided substantial contributions to the conception of the work, acquisition and interpretation of the data, particularly for the mass spectrometry setup and the refinement of the targeted panel. S. Schade, S.W., C.T., M.B., B.M., M.-L.M. and F.S.D. conceptualised the clinical study, analyzed the clinical data and reevaluated the diagnosis. M.E. provided substantial contributions to the clinical data analyzes, particularly the imaging patient data in regard to differential diagnosis. J.H., M.B., K.M. and B.M. wrote the manuscript with input and substantial revisions from all authors.
Corresponding authors
Correspondence to Jenny Hällqvist or Michael Bartl .
Ethics declarations
Competing interests.
JH, MD, MX, SW, KB, ME, PG, MGB, CP, KM, ID, WH, JS, HV and CF and have no competing interests to report. MB has received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – 413,501,650. CT has received honoraria for consultancy from Roche, and honoraria for educational lectures from UCB, and has received research funding for the PPMI study from the Michael J. Fox Foundation and funding from the EU (Horizon 2020) and stipends from the (International Parkinson’s and Movement Disorder Society) IPMDS. BM has received honoraria for consultancy from Roche, Biogen, AbbVie, UCB, and Sun Pharma Advanced Research Company. BM is a member of the executive steering committee of the Parkinson Progression Marker Initiative and PI of the Systemic Synuclein Sampling Study of the Michael J. Fox Foundation for Parkinson’s Research and has received research funding from the Deutsche Forschungsgemeinschaft (DFG), EU (Horizon 2020), Parkinson Fonds Deutschland, Deutsche Parkinson Vereinigung, Parkinson’s Foundation and the Michael J. Fox Foundation for Parkinson’s Research. MLM has received honoraria for speaking engagements from Deutsche Parkinson Gesellschaft e.V., and royalties from Gesellschaft fur Medien + Kommunikation mbH + Co. FSD has received honoraria for speaking engagements from AbbVie, Bial, Ever Pharma, Medtronic and royalties from Elsevier and Springer. She served on an advisory board for Zambon and Stada Pharma. FSD participated in Ad Boards for consultation: Abbvie, UCB, Bial, Ono, Roche and got honorary for lecturing: Stada Pharm, AbbVie, Alexion, Bial. S. Schade received institutional salaries supported by the EU Horizon 2020 research and innovation programme under grant agreement No. 863664 and by the Michael J. Fox Foundation for Parkinson’s Research under grant agreement No. MJFF-021923. He is supported by a PPMI Early Stage Investigators Funding Programme fellowship of the Michael J. Fox Foundation for Parkinson’s Research under grant agreement No. MJFF-022656. S. Schreglmann received institutional salaries supported by the EU Horizon 2020 research and innovation programme under grant agreement No. 863664, support from the Advanced Clinician Scientist programme by the Interdisciplinary Centre for Clinical Research, Wuerzburg, Germany, and from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Project-ID 424778381-TRR 295. He is a fellow of the Thiemann Foundation. He serves as a scientific adviser to Elemind Inc.
Peer review
Peer review information.
Nature Communications thanks Kanta Horie and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Hällqvist, J., Bartl, M., Dakna, M. et al. Plasma proteomics identify biomarkers predicting Parkinson’s disease up to 7 years before symptom onset. Nat Commun 15 , 4759 (2024). https://doi.org/10.1038/s41467-024-48961-3
Download citation
Received : 06 April 2023
Accepted : 20 May 2024
Published : 18 June 2024
DOI : https://doi.org/10.1038/s41467-024-48961-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

Will China’s historic paper on Chang’e-6 lunar far side samples be in English or Chinese?
- The Chang’e-6 mission’s cargo is expected to yield a wealth of research but debate is growing about what language it will be published in first

Within China’s scientific community, there persists a notion that publishing in English is not only a medium of communication, but also a bridge to global recognition. The use of Chinese remains taboo, a silent sacrifice at the altar of international acceptance.
When China’s Chang’e-5 mission in 2020 retrieved the first lunar samples in decades from the moon’s near side, the first research was carried out by a joint team of Chinese and Western scientists and appeared in Science magazine in October 2021.
This was followed by three more scientific papers published by Nature in the same month, according to an editor with the Science China Press, a scientific journal publishing company of the Chinese Academy of Sciences (CAS), recalling the global sensation.
The rocks collected in 2020 led to a number of surprising discoveries, as they turned out to be much younger than the samples brought back by the US Apollo and Soviet Luna missions in the 1960s and 1970s.
“We certainly hope that some of our country’s groundbreaking scientific and technological achievements can appear in China’s top journals, so that we can expand our influence,” said the editor, who asked not to be named.
It was not always so. Tu Youyou, who won China’s first Nobel Prize for science in 2015, published her paper on the discovery of artemisinin in the Chinese Science Bulletin in 1977.
The journal, co-sponsored by CAS and the National Natural Science Foundation of China, once published many major discoveries but since the 1990s has suffered from a lack of quality manuscripts.

Speaking at a conference in 2018, George Gao Fu – a leading scientist in the field of virology and immunology and former head of the Chinese Centre for Disease Control and Prevention (CDC) – said Chinese as a language of academic communication “used to be glorious”.
Breakthroughs, including Tu’s achievement and the discovery of high-temperature iron-based superconducting materials, had been published first in Chinese-language journals and then recognised by the world, he said.
However, for three decades China’s important scientific research results were “basically first reported by foreign journals”, Gao noted.
Interestingly, just a few years later, Gao led a landmark study by a Chinese CDC team on the epidemiology of Covid-19 that was first published in January 2020 by the New England Journal of Medicine.
The move caused controversy in China, where the public was eager for any information about the new coronavirus that causes Covid-19 as the country grappled with the early stages of the pandemic.
The response to the overseas publication of the study reflected a broader, uncomfortable dilemma for Chinese researchers: while they recognise the importance or necessity of writing in their native language, it is difficult on a practical level.
China’s Chang’e-6 touches down on far side of moon on mission to bring rock samples back to Earth
Newton’s Principia Mathematica was written in Latin. Einstein’s first influential papers were written in German. Marie Curie’s work was published in French.
Yet, since the middle of the last century, there has been a shift in the global scientific community, with most scientific research now published in a single language – English, which is spoken by only about 18 per cent of the world’s population.
While it is estimated that up to 98 per cent of global scientific research is published in English, the number of papers by Chinese scholars has been climbing.
As early as 2010, Chinese biologist Zhu Zuoyan, a CAS academician, observed that the number of papers published by scholars from China had risen from 0.2 per cent of the world’s total to 10 per cent within a decade, second only to the United States.
But China’s academic evaluation system encourages the flow of excellent papers to foreign journals, which had partly led to the country’s lack of international academic impact, despite having the second largest number of academic journals – more than 4,800 – in the world, he said.
In late 2019, Li Zhimin, former director of the Ministry of Education’s Science and Technology Development Centre, called for papers to be published in the country’s official language if the research is funded by the government.
The requirement would make it easier for funders to review research projects, facilitate exchanges with their domestic counterparts and improve the nation’s scientific literacy, he said.
A CAS physicist, who declined to be named, stressed that the proposal to “write research results on the soil of the motherland” could not simply be understood as submitting and publishing articles in domestic journals and in Chinese.
That would be “parochial”, he said. Rather, the key is to focus research on solving crucial issues or problems in China’s development, rather than blindly following global research hotspots and wasting research funds and resources.
But at an individual level, there are plenty of pragmatic reasons and incentives for researchers to do just that. Under China’s evaluation system, getting articles published in prestigious English-language journals often brings rewards.
In addition to promotion opportunities and academic honours, there is also fame, with overseas scientific recognition tending to attract wide media and public attention.
Last month, for example, biologist Zhu Jiapeng earned a prize from Nanjing University of Traditional Chinese Medicine for his “outstanding contribution” and a grant of 1 million yuan (US$138,000) as one of the lead authors in a study published by Nature.
Astronomer Deng Licai, with the National Astronomical Observatories under CAS, believes that research results from national missions such as the Chang’e programme should be prioritised for publication in domestic journals.
Deng, who has been a team leader on China’s giant telescope project since its development in the 1990s and 2000s, said he insisted that the first batch of studies to emerge from the Large Sky Area Multi-Object Fibre Spectroscopic Telescope (Lamost) appeared in domestic journals.
“This can firstly highlight the nationality of these independent and cutting-edge major scientific projects, and also help to enhance the international impact of domestic academic journals,” he said.
But English has become the international scientific community’s lingua franca and should be used as a medium of communication, Deng said, adding that it had nothing to do with politics.
China’s space plans: lunar GPS, a 3D-printed moon base and soil samples from Mars
According to Deng, the scientists who study the Chang’e-6 lunar samples could consider publishing in some of China’s English-language journals, such as Research in Astronomy and Astrophysics (RAA).
“All of our pre-research articles on the Lamost programme published in RAA have made it into international lists of highly cited articles,” he said.
Chinese Academy of Social Sciences researcher Zhu Rui, who prefers to publish in Chinese journals – partly because the academy encourages it – said that using his own language when writing academic papers is not an obstacle, as long as the scientific community maintains substantive communication.
| Acta Crystallographica Section D Acta Crystallographica Section D STRUCTURAL BIOLOGY |
1. Introduction
4. discussion, 5. related literature, supporting information.

| Format | BIBTeX | |
| EndNote | ||
| RefMan | ||
| Refer | ||
| Medline | ||
| CIF | ||
| SGML | ||
| Plain Text | ||
| Text | ||

research papers \(\def\hfill{\hskip 5em}\def\hfil{\hskip 3em}\def\eqno#1{\hfil {#1}}\)
| STRUCTURAL BIOLOGY |

Validation of electron-microscopy maps using solution small-angle X-ray scattering
a Department of Chemistry and Interdisciplinary Nanoscience Center (iNANO), Aarhus University, Gustav Wieds Vej 14, 8000 Aarhus, Denmark * Correspondence e-mail: [email protected]
The determination of the atomic resolution structure of biomacromolecules is essential for understanding details of their function. Traditionally, such a structure determination has been performed with crystallographic or nuclear resonance methods, but during the last decade, cryogenic transmission electron microscopy (cryo-TEM) has become an equally important tool. As the blotting and flash-freezing of the samples can induce conformational changes, external validation tools are required to ensure that the vitrified samples are representative of the solution. Although many validation tools have already been developed, most of them rely on fully resolved atomic models, which prevents early screening of the cryo-TEM maps. Here, a novel and automated method for performing such a validation utilizing small-angle X-ray scattering measurements, publicly available through the new software package AUSAXS , is introduced and implemented. The method has been tested on both simulated and experimental data, where it was shown to work remarkably well as a validation tool. The method provides a dummy atomic model derived from the EM map which best represents the solution structure.
Keywords: electron microscopy ; small-angle X-ray scattering ; electron-microscopy validation ; structure determination .
Small-angle X-ray scattering (SAXS) is an alternative but low-resolution technique for structural analysis. While similar in principle to X-ray crystallography based on interference of scattered X-rays, the requirement for crystallization is evaded by simultaneously measuring the scattering pattern of multiple molecules in solution. The result is a single one-dimensional orientationally averaged intensity curve that is dependent on both the shape and size of the sample. One of the primary advantages of SAXS is that macromolecular molecules and complexes can be measured in their native state in solution, without any special sample preparation. This feature is exactly what makes the technique so useful for validation.
The next section will present and detail the method itself, including brief discussions of all of the major design decisions. This is followed by a section detailing how the method has been tested with both simulated and experimental data, along with tables of all test results.
In TEM, the electrons interact with the electric field generated by the individual atoms of the sample molecule. Since these fields are continuous, the surface of the molecule is not well defined in an EM map. When visualizing such a map with, for example, PyMOL , one must instead pick some threshold cutoff value, which is then used to define the surface. A 3D TEM map thus represent the Coulomb charge -density distribution, represented on a grid with a resolution dependent on the experimental setup.
| A helpful way of visualizing EM maps. The left panel shows the typical visualization from . The right panel shows how the maps can also be interpreted as a stack of 2D contour plots. |
2.1. Model generation
The creation of a single model for some threshold value thus involves first placing weighted (either by density or by some constant) dummy atoms and then simulating a hydration layer. The next step is to vary this threshold value to generate an entire series of dummy models of varying sizes. Note that the models for nearby threshold values are expected to be very similar.
2.2. Model selection
Although there are already plenty of programs that can calculate these expected scattering curves for the models ( CRYSOL , FoXS , …), we decided to use our own implementation. There are two primary reasons for this choice.
| , 2013 ), by adding layers of uniformly distributed electron density around the surface (Svergun , 1995 ; Grudinin , 2017 ), or with explicit molecular-dynamics simulation calculations (Knight & Hub, 2015 ). Since performing actual simulations is too slow for our purposes, we believe the best alternative is to actually model the hydration molecules as randomly distributed dummy solvent atoms close to the protein surface. into the Debye equation, a major performance improvement can be achieved when calculating the total histograms and expected scattering curves of similar structures. The idea is that by splitting the EM map into an onion-like structure with regions of similar density values, it becomes possible to reuse previous scattering calculations when scanning the threshold value. More specifically, the threshold value is scanned from its highest value to its lowest value while saving the self-correlation histogram of each `onion shell'. The self-correlations from the inner shells can then directly be reused when evaluating the scattering from a threshold value outside their region. Thus, instead of being a ( ) process in the number of atoms, evaluating the scattering from similar structures is improved to an ( ) process, where is the number of additional scatterers. With the threshold parameter being nearly continuous and by scanning from high to low, thus creating a series of similar models, is small compared with . Implementing optimizations such as this in existing libraries is a major undertaking, and is impossible for the closed-source . Developing a new library that natively supports these partial histograms was the easiest solution. We will return to this performance discussion later. |
For highly ordered structures, such as the lattice structure of the maps, it turned out that using the binned distance approximation typically used in conjunction with the Debye equation resulted in significant inaccuracies. This is because in such highly ordered structures some distances are much likelier than others, yet the binning does not account for this and shifts them to the centre of the closest bin. With almost every single distance being shifted by a small amount, the error propagates into a significant uncertainty in the final scattering profile. To solve this issue, we introduced weighted bins into the approximation, where the centre of the bin is determined based on its contents, calculated as the centre of mass of the bin. This neatly solves the issue, while still providing the significant performance benefit of the binning approximation. Note that using weighted bins is usually not necessary when evaluating the scattering of a typical protein, only when dealing with highly ordered structures, as we are here.
The method optimizes four parameters in total, where the first is the threshold value itself. As explained previously, for efficiency reasons this parameter is scanned using a fixed step size, starting from its highest value and moving towards its lowest, thus generating a number of equidistant dummy models. For each of these models, three additional parameters are optimized: two for the simple linear fit to the scattering data I exp = aI model + b , and a third for fitting the scattering contrast of the hydration layer. Although adding the hydration layer generally provides a dramatic improvement to χ 2 , it also comes with a major drawback: the scattering contrast parameter is strongly correlated to the threshold value. This is only to be expected, as they both control the effective size of the model: the former by enhancing the scattering contribution from the dummy water surface atoms and the latter by directly varying the size. The strong correlation between these parameters naturally leads to large uncertainties in them, although this is not a concern as the former is an arbitrary scaling constant and the latter is only approximative. What is more problematic is the discrete nature of the data stored in the maps, with a small but finite difference between the density values of neighbouring voxels. When the threshold value crosses such a boundary, a number of new dummy atoms are added to the model proportional to the current surface area , while the total number of dummy atoms is of course proportional to the volume . Thus, for small volumes the scattering contribution of the newly added dummy atoms is significant, leading to a high variance in this region of the χ 2 landscape. Typically, the extreme low-volume region is not of interest for the fit itself, meaning that only limited variance is observed in the relevant area of the landscape. The problem is further mitigated by using a moving average as an estimate of the actual χ 2 .
Since the threshold value is directly related to the size of the dummy model, there is in principle a one-to-one mapping from the threshold value to the total mass of the model. With this mapping the threshold axis can be replaced with a mass axis, which may be useful for real applications, especially in cases with multiple minima in the χ 2 landscape. Since dummy models are generated for all identified minima, the user can then subsequently select only the one that they are interested in based on the mass. It should be mentioned that this mass axis comes with a significant uncertainty and may be unsuitable for absolute comparisons.
The ideal test would be to simulate both SAXS scattering curves and EM maps from a complete atomic structure, while also varying the resolution of the map. While the former is doable, the latter is a nontrivial problem that currently only has approximate solutions. This immediately makes this approach unusable, since one cannot determine whether a bad fit is due to issues with the map simulation or due to the method itself. We have thus focused on simulating SAXS data for our EM tests.
To better emulate experimental data, each point of the scattering curve should have an error associated with it. By comparison to a series of measured SAXS data sets, we have empirically found the errors to be reasonably well described by the equation
After the errors have been calculated using this equation, Gaussian noise with this magnitude is imposed on the simulated data.
3.1. Examples using experimental EM maps and simulated SAXS data
As part of the standard validation suites required before deposition of an EM map, a high-resolution atomic structure model is built and refined to fit the map itself. Since this fitted structure should be a good representation of the map, it can be used for testing, i.e. we can use the high-resolution model to generate a simulated SAXS data set for the test. We would then expect the agreement to be good, but not necessarily perfect. The tests will also serve as guidelines for the kind of results and agreements that one can expect from the method in general.
A random selection of maps covering a wide range of resolutions was downloaded for this test. SAXS measurements were then simulated for each as described above, and subsequently fitted by the scattering from the map itself as per the method described in the present paper, using unity weights.
IMAGES
VIDEO
COMMENTS
Table of contents. What not to include in your discussion section. Step 1: Summarize your key findings. Step 2: Give your interpretations. Step 3: Discuss the implications. Step 4: Acknowledge the limitations. Step 5: Share your recommendations. Discussion section example. Other interesting articles.
The discussion section is one of the final parts of a research paper, in which an author describes, analyzes, and interprets their findings. They explain the significance of those results and tie everything back to the research question(s). In this handout, you will find a description of what a discussion section does, explanations of how to ...
Begin with a clear statement of the principal findings. This will reinforce the main take-away for the reader and set up the rest of the discussion. Explain why the outcomes of your study are important to the reader. Discuss the implications of your findings realistically based on previous literature, highlighting both the strengths and ...
1.Introduction—mention gaps in previous research¹⁻². 2. Summarizing key findings—let your data speak¹⁻². 3. Interpreting results—compare with other papers¹⁻². 4. Addressing limitations—their potential impact on the results¹⁻². 5. Implications for future research—how to explore further¹⁻².
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research. This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study ...
Papers usually end with a concluding section, often called the "Discussion.". The Discussion is your opportunity to evaluate and interpret the results of your study or paper, draw inferences and conclusions from it, and communicate its contributions to science and/or society. Use the present tense when writing the Discussion section.
An example of research summary in discussion. 3.2. An example of result interpretation in discussion. 3.3. An example of literature comparison in discussion. 3.4. An example of research implications in discussion. 3.5. An example of limitations in discussion.
The discussion section is often considered the most important part of your research paper because it: Most effectively demonstrates your ability as a researcher to think critically about an issue, to develop creative solutions to problems based upon a logical synthesis of the findings, and to formulate a deeper, more profound understanding of the research problem under investigation;
Step 1: Restate your research problem and research questions. The first step in writing up your discussion chapter is to remind your reader of your research problem, as well as your research aim (s) and research questions. If you have hypotheses, you can also briefly mention these.
Begin the Discussion section by restating your statement of the problem and briefly summarizing the major results. Do not simply repeat your findings. Rather, try to create a concise statement of the main results that directly answer the central research question that you stated in the Introduction section.
Papers that are submitted to a journal for publication are sent out to several scientists (peers) who look carefully at the paper to see if it is "good science". These reviewers then recommend to the editor of a journal whether or not a paper should be published. Most journals have publication guidelines. Ask for them and follow them exactly.
In an empirical research paper, the purpose of the Discussion section is to interpret the results and discuss their implications, thereby establishing (and often qualifying) the practical and scholarly significance of the present study. It may be helpful to think of the Discussion section as the inverse of the introduction to an empirical ...
Tips to Write the Results Section. Direct the reader to the research data and explain the meaning of the data. Avoid using a repetitive sentence structure to explain a new set of data. Write and highlight important findings in your results. Use the same order as the subheadings of the methods section.
Discussion Section. The overall purpose of a research paper's discussion section is to evaluate and interpret results, while explaining both the implications and limitations of your findings. Per APA (2020) guidelines, this section requires you to "examine, interpret, and qualify the results and draw inferences and conclusions from them ...
Discussion is mainly the section in a research paper that makes the readers understand the exact meaning of the results achieved in a study by exploring the significant points of the research, its ...
Step 3: Relate to Existing Literature. In this step, link up your discoveries with what other researchers have already figured out. Share if your results are similar to or different from what's been found before. This helps give more background to your study and shows you know what other scientists have been up to.
The discussion section can be written in 3 parts: an introductory paragraph, intermediate paragraphs and a conclusion paragraph. For intermediate paragraphs, a "divide and conquer" approach, meaning a full paragraph describing each of the study endpoints, can be used. In conclusion, academic writing is similar to other skills, and practice ...
The discussion section, a systematic critical appraisal of results, is a key part of a research paper, wherein the authors define, critically examine, describe and interpret their findings ...
The discussion section of a research paper is where the author analyzes and explains the importance of the study's results. It presents the conclusions drawn from the study, compares them to previous research, and addresses any potential limitations or weaknesses. The discussion section should also suggest areas for future research.
Writing a discussion section is where you really begin to add your interpretations to the work. In this critical part of the research paper, you start the process of explaining any links and correlations apparent in your data. If you left few interesting leads and open questions in the results section, the discussion is simply a matter of ...
1. Begin by discussing the research question and talking about whether it was answered in the research paper based on the results. 2. Highlight any unexpected and/or exciting results and link them to the research question. 3. Point out some previous studies and draw comparisons on how your study is different. 4.
This template covers all the core components required in the discussion/analysis chapter of a typical dissertation or thesis, including: The purpose of each section is explained in plain language, followed by an overview of the key elements that you need to cover. The template also includes practical examples to help you understand exactly what ...
Future research on this topic would benefit from the inclusion of control samples (plants or plant parts not consumed by chimpanzees); however, in this study, assay costs were a prohibiting factor. Additional information regarding the nutritional and mineral content of the species mentioned in this study is needed to better understand the ...
Table of contents. What not to include in your discussion section. Step 1: Summarise your key findings. Step 2: Give your interpretations. Step 3: Discuss the implications. Step 4: Acknowledge the limitations. Step 5: Share your recommendations. Discussion section example.
Discussions of Previously Published Papers. Discussion on "Sample Size Determination for Credibility Estimation," by Liang Hong, Volume 26(4) ... Register to receive personalised research and resources by email. Sign me up. Taylor and Francis Group Facebook page.
Schreglmann received institutional salaries supported by the EU Horizon 2020 research and innovation programme under grant agreement No. 863664, support from the Advanced Clinician Scientist ...
Discussions of Previously Published Papers. Author's Reply to Discussion on "Sample Size Determination for Credibility Estimation" ... Register to receive personalised research and resources by email. Sign me up. Taylor and Francis Group Facebook page.
When China's Chang'e-5 mission in 2020 retrieved the first lunar samples in decades from the moon's near side, the first research was carried out by a joint team of Chinese and Western ...
2.1. Model generation. The first task is to construct a set of dummy-atom models for an EM map. The simplest way of constructing such a model is by imposing a threshold cutoff value d, similar to the alpha level in PyMOL, below which the density is assumed to be noise and is therefore discarded.The intrinsic grid of the map itself is then used to place dummy atoms, each weighted either by the ...