Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Review Article
- Published: 10 November 2015

Methods and models for unravelling human evolutionary history
- Joshua G. Schraiber 1 &
- Joshua M. Akey 1
Nature Reviews Genetics volume 16 , pages 727–740 ( 2015 ) Cite this article
15k Accesses
89 Citations
32 Altmetric
Metrics details
- Evolutionary biology
- Evolutionary genetics
- Genetic variation
- Next-generation sequencing
- Statistical methods
High-throughput sequencing is enabling massively large catalogues of DNA sequence variation to be collected in geographically diverse human populations. Such data sets contain considerable information about human history but are complex and require careful analysis.
Quality control and exploratory data analyses are critical in analyses of large-scale sequencing data sets and help to identify features of the data that may complicate downstream inferences.
Functional and comparative genomics data (such as sequence conservation, chromatin immunoprecipitation followed by sequencing (ChIP–seq) and DNase I hypersensitivity) can be leveraged to mitigate the confounding effect of natural selection when inferring demographic models.
A large number of flexible and sophisticated methods have been developed that allow specific and detailed demographic inferences to be made. The appropriate method to use depends on the specific hypothesis or question being asked, and the underlying assumptions of a given method should be carefully considered.
As sample sizes become increasingly large, inferences about specific aspects of breeding structure and demography may be possible. However, these methods are still in their infancy and require substantial theoretical and methodological development.
The increasing availability of ancient DNA from modern and archaic humans provides exciting new possibilities to refine parameters of human evolutionary history, although new methodological development is needed to fully realize the potential of these data.
The genomes of contemporary humans contain considerable information about the history of our species. Although the general contours of human evolutionary history have been defined with increasing resolution throughout the past several decades, the continuing deluge of massively large sequencing data sets presents new opportunities and challenges for understanding human evolutionary history. Here, we review the signatures that demographic history imparts on patterns of DNA sequence variation, statistical methods that have been developed to leverage information contained in genome-scale data sets and insights gleaned from these studies. We also discuss the importance of using exploratory analyses to assess data quality, the strengths and limitations of commonly used population genomics methods, and factors that confound population genomics inferences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
176,64 € per year
only 14,72 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others

Estimation of coalescence probabilities and population divergence times from SNP data

A method for genome-wide genealogy estimation for thousands of samples
Incongruence in the phylogenomics era
Veeramah, K. R. & Hammer, M. F. The impact of whole-genome sequencing on the reconstruction of human population history. Nat. Rev. Genet. 15 , 149–162 (2014).
CAS PubMed Google Scholar
Metzker, M. L. Sequencing technologies — the next generation. Nat. Rev. Genet. 11 , 31–46 (2010).
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491 , 56–65 (2012). This study describes an international project that created one of the most-comprehensive catalogues of sequence variation in geographically diverse populations.
Tennessen, J. A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337 , 64–69 (2012). This article represents one of the earliest large-scale, high-coverage exome data sets to be produced; it has been extensively used in evolutionary and medical genomics.
CAS PubMed PubMed Central Google Scholar
Bustamante, C. D., De La Vega, F. M. & Burchard, E. G. Genomics for the world. Nature 475 , 163–165 (2011).
Gravel, S. et al. Demographic history and rare allele sharing among human populations. Proc. Natl Acad. Sci. USA 108 , 11983–11988 (2011).
Hellenthal, G. et al. A genetic atlas of human admixture history. Science 343 , 747–751 (2014).
Leslie, S. et al. The fine-scale genetic structure of the British population. Nature 519 , 309–314 (2015).
Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 39 , 197–218 (2005).
Sabeti, P. C. et al. Positive natural selection in the human lineage. Science 312 , 1614–1620 (2006).
Bamshad, M. & Wooding, S. P. Signatures of natural selection in the human genome. Nat. Rev. Genet. 4 , 99–111 (2003).
Akey, J. M. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 19 , 711–722 (2009).
Fu, W. & Akey, J. M. Selection and adaptation in the human genome. Annu. Rev. Genom. Hum. Genet. 14 , 467–489 (2013).
CAS Google Scholar
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20 , 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43 , 491–498 (2011).
Auwera, G. A. et al. From fastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 15 , 1110 (2013).
Google Scholar
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15 , 356 (2014).
PubMed PubMed Central Google Scholar
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 , 1851–1858 (2008).
Schraiber, J. G., Shih, S. & Slatkin, M. Genomic tests of variation in inbreeding among individuals and among chromosomes. Genetics 192 , 1477–1482 (2012).
Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 41 , 1061–1067 (2009).
Williamson, S. H. et al. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl Acad. Sci. USA 102 , 7882–7887 (2005). This study reports a clever approach to account for the effects of selection when making demographic inferences.
Živkovic, D., Steinrücken, M., Song, Y. S. & Stephan, W. Transition densities and sample frequency spectra of diffusion processes with selection and variable population size. Genetics 200 , 601–617 (2015).
Hammer, M. F. et al. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat. Genet. 42 , 830–831 (2010).
Gottipati, S., Arbiza, L., Siepel, A., Clark, A. G. & Keinan, A. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat. Genet. 43 , 741–743 (2011).
Gazave, E. et al. Neutral genomic regions refine models of recent rapid human population growth. Proc. Natl Acad. Sci. USA 111 , 757–762 (2014). This study illustrates well how choosing neutral genomic regions carefully can lead to more-refined estimates of demographic parameters.
Consortium, T. E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489 , 57–74 (2012).
Romanoski, C. E., Glass, C. K., Stunnenberg, H. G., Wilson, L. & Almouzni, G. Epigenomics: roadmap for regulation. Nature 518 , 314–316 (2015).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20 , 110–121 (2010).
McVicker, G., Gordon, D., Davis, C. & Green, P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5 , e1000471 (2009).
Pollard, K. S. et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2 , e168 (2006).
Arbiza, L., Zhong, E. & Keinan, A. NRE: a tool for exploring neutral loci in the human genome. BMC Bioinformatics 13 , 301 (2012).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155 , 945–959 (2000). This classic paper describes a nonparametric approach for inferring population structure.
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The History And Geography Of Human Genes (Princeton Univ. Press, 1994).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456 , 98–101 (2008).
Biswas, S., Scheinfeldt, L. B. & Akey, J. M. Genome-wide insights into the patterns and determinants of fine-scale population structure in humans. Am. J. Hum. Genet. 84 , 641–650 (2009).
McVean, G. A. Genealogical interpretation of principal components analysis. PLoS Genet. 5 , e1000686 (2009).
François, O. et al. Principal component analysis under population genetic models of range expansion and admixture. Mol. Biol. Evol. 27 , 1257–1268 (2010).
PubMed Google Scholar
Novembre, J. & Stephens, M. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 40 , 646–649 (2008).
Yang, W.-Y., Novembre, J., Eskin, E. & Halperin, E. A model-based approach for analysis of spatial structure in genetic data. Nat. Genet. 44 , 725–731 (2012).
Tang, H., Peng, J., Wang, P. & Risch, N. J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28 , 289–301 (2005).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19 , 1655–1664 (2009).
Raj, A., Stephens, M. & Pritchard, J. K. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197 , 573–589 (2014).
Huelsenbeck, J. P. & Andolfatto, P. Inference of population structure under a Dirichlet process model. Genetics 175 , 1787–1802 (2007).
Xie, W., Lewis, P. O., Fan, Y., Kuo, L. & Chen, M.-H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 60 , 150–160 (2010).
Patterson, N. et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 74 , 979–1000 (2004).
Gravel, S. Population genetics models of local ancestry. Genetics 191 , 607–619 (2012).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165 , 2213–2233 (2003).
Price, A. L. et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5 , e1000519 (2009).
Lawson, D. J., Hellenthal, G., Myers, S. & Falush, D. Inference of population structure using dense haplotype data. PLoS Genet. 8 , e1002453 (2012).
Pool, J. E. & Nielsen, R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181 , 711–719 (2009).
Liang, M. & Nielsen, R. The lengths of admixture tracts. Genetics 197 , 953–967 (2014).
Sankararaman, S., Sridhar, S., Kimmel, G. & Halperin, E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 82 , 290–303 (2008).
Brisbin, A. et al. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum. Biol. 84 , 343–364 (2012).
Wakeley, J. Coalescent Theory: An Introduction (Robert & Co., 2009).
Sawyer, S. A. & Hartl, D. L. Population genetics of polymorphism and divergence. Genetics 132 , 1161–1176 (1992).
Bhaskar, A. & Song, Y. S. Descartes' rule of signs and the identifiability of population demographic models from genomic variation data. Ann. Statist. 42 , 2469–2493 (2014).
Terhorst, J. & Song, Y. S. Fundamental limits on the accuracy of demographic inference based on the sample frequency spectrum. Proc. Natl Acad. Sci. USA 112 , 7677–7682 (2015).
Bustamante, C. D., Wakeley, J., Sawyer, S. & Hartl, D. L. Directional selection and the site-frequency spectrum. Genetics 159 , 1779–1788 (2001).
Evans, S. N., Shvets, Y. & Slatkin, M. Non-equilibrium theory of the allele frequency spectrum. Theor. Popul. Biol. 71 , 109–119 (2007).
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5 , e1000695 (2009).
Lukic, S. & Hey, J. Demographic inference using spectral methods on SNP data, with an analysis of the human out-of-Africa expansion. Genetics 192 , 619–639 (2012).
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., Sousa, V. C. & Foll, M. Robust demographic inference from genomic and SNP data. PLoS Genet. 9 , e1003905 (2013).
Excoffier, L. & Foll, M. Fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27 , 1332–1334 (2011).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8 , e1002967 (2012).
Bhaskar, A., Wang, Y. & Song, Y. S. Efficient inference of population size histories and locus-specific mutation rates from large-sample genomic variation data. Genome Res. 25 , 268–279 (2014).
Griffiths, R. C. & Marjoram, P. An ancestral recombination graph. University of Canterbury [online] , (1997).
Wiuf, C. & Hein, J. Recombination as a point process along sequences. Theor. Popul. Biol. 55 , 248–259 (1999).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81 , 1084–1097 (2007).
Gusev, A. et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 19 , 318–326 (2009).
Ralph, P. & Coop, G. The geography of recent genetic ancestry across Europe. PLoS Biol. 11 , e1001555 (2013).
Palamara, P. F., Lencz, T., Darvasi, A. & Pe'er, I. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 91 , 809–822 (2012).
Palamara, P. F. & Pe'er, I. Inference of historical migration rates via haplotype sharing. Bioinformatics 29 , i180–i188 (2013).
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194 , 459–471 (2013).
McVean, G. A. T. & Cardin, N. J. Approximating the coalescent with recombination. Philos. Trans. R. Soc. B Biol. Sci. 360 , 1387–1393 (2005). This article introduces the SMC, which enabled important developments in population genomic inferencing from recombining sequences.
Marjoram, P. & Wall, J. D. Fast 'coalescent' simulation. BMC Genet. 7 , 16 (2006).
Harris, K. & Nielsen, R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 9 , e1003521 (2013).
Liu, S. et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157 , 785–794 (2014).
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475 , 493–496 (2011). This study describes PSMC, which enables quasi-non-parametric inferencing of effective population size through time from a single diploid genome sequence.
Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22 , 1185–1192 (2005).
Heled, J. & Drummond, A. J. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8 , 289 (2008). This study details one of the first, and underappreciated, methods to infer population size history in a relatively non-parametric way from haplotype data.
Minin, V. N., Bloomquist, E. W. & Suchard, M. A. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25 , 1459–1471 (2008).
Schiffels, S. & Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46 , 919–925 (2014).
Hobolth, A., Christensen, O. F., Mailund, T. & Schierup, M. H. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3 , e7 (2007).
Dutheil, J. Y. et al. Ancestral population genomics: the coalescent hidden Markov model approach. Genetics 183 , 259–274 (2009).
Mailund, T., Dutheil, J. Y., Hobolth, A., Lunter, G. & Schierup, M. H. Estimating divergence time and ancestral effective population size of bornean and sumatran orangutan subspecies using a coalescent hidden Markov model. PLoS Genet. 7 , e1001319 (2011).
Hobolth, A., Dutheil, J. Y., Hawks, J., Schierup, M. H. & Mailund, T. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 21 , 349–356 (2011).
Mailund, T. et al. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet. 8 , e1003125 (2012).
Scally, A. et al. Insights into hominid evolution from the gorilla genome sequence. Nature 483 , 169–175 (2012).
Sheehan, S., Harris, K. & Song, Y. S. Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194 , 647–662 (2013).
Stephens, M. & Donnelly, P. Inference in molecular population genetics. J. R. Stat. Soc. B 62 , 605–655 (2000).
Paul, J. S., Steinrücken, M. & Song, Y. S. An accurate sequentially Markov conditional sampling distribution for the coalescent with recombination. Genetics 187 , 1115–1128 (2011).
Steinrücken, M., Paul, J. S. & Song, Y. S. A sequentially Markov conditional sampling distribution for structured populations with migration and recombination. Theor. Popul. Biol. 87 , 51–61 (2013).
Kuhner, M. K. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22 , 768–770 (2006).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29 , 1969–1973 (2012).
Rannala, B. & Yang, Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164 , 1645–1656 (2003).
Gronau, I., Hubisz, M. J., Gulko, B., Danko, C. G. & Siepel, A. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43 , 1031–1034 (2011).
Lohse, K., Harrison, R. J. & Barton, N. H. A general method for calculating likelihoods under the coalescent process. Genetics 189 , 977–987 (2011).
Lohse, K. & Frantz, L. A. F. Neandertal admixture in Eurasia confirmed by maximum-likelihood analysis of three genomes. Genetics 196 , 1241–1251 (2014).
Rasmussen, M. D., Hubisz, M. J., Gronau, I. & Siepel, A. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 10 , e1004342 (2014).
Ségurel, L., Wyman, M. J. & Przeworski, M. Determinants of mutation rate variation in the human germline. Annu. Rev. Genomics Hum. Genet. 15 , 47–70 (2014). This review covers in great detail the recent controversy about the human genomic mutation rate and summarizes the different kinds of mutations in the human genome.
Bhaskar, A., Clark, A. G. & Song, Y. S. Distortion of genealogical properties when the sample is very large. Proc. Natl Acad. Sci. USA 111 , 2385–2390 (2014).
Wakeley, J., King, L., Low, B. S. & Ramachandran, S. Gene genealogies within a fixed pedigree, and the robustness of Kingman's coalescent. Genetics 190 , 1433–1445 (2012).
Möhle, M. Robustness results for the coalescent. J. Appl. Probab. 35 , 438–447 (1998). This important theory paper outlines the broad generality of the Kingman coalescent.
Pitman, J. Coalescents with multiple collisions. Ann. Appl. Probab. 27 , 1870–1902 (1999).
Sagitov, S. The general coalescent with asynchronous mergers of ancestral lines. J. Appl. Probab. 36 , 1116–1125 (1999).
Zerjal, T. et al. The genetic legacy of the Mongols. Am. J. Hum. Genet. 72 , 717–721 (2003).
Varin, C., Reid, N. & Firth, D. An overview of composite likelihood methods. Statist. Sin. 21 , 5–42 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 , 559–575 (2007).
Beaumont, M. A. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41 , 379–406 (2010).
Beaumont, M. A., Zhang, W. & Balding, D. J. Approximate Bayesian computation in population genetics. Genetics 162 , 2025–2035 (2002).
Sunnåker, M. et al. Approximate Bayesian computation. PLoS Comput. Biol. 9 , e1002803 (2013).
Csilléry, K., Blum, M. G. B., Gaggiotti, O. E. & François, O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 25 , 410–418 (2010).
Wegmann, D., Leuenberger, C. & Excoffier, L. Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics 182 , 1207–1218 (2009).
Sisson, S. A., Fan, Y. & Tanaka, M. M. Sequential Monte Carlo without likelihoods. Proc. Natl Acad. Sci. USA 104 , 1760–1765 (2007).
Wegmann, D., Leuenberger, C., Neuenschwander, S. & Excoffier, L. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11 , 116 (2010).
Fearnhead, P. & Prangle, D. Constructing summary statistics for approximate Bayesian computation: semi-automatic approximate Bayesian computation. J. R. Stat. Soc. 74 , 419–474 (2012).
Pickrell, J. K. & Reich, D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 30 , 377–389 (2014).
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328 , 710–722 (2010).
Plagnol, V. & Wall, J. D. Possible ancestral structure in human populations. PLoS Genet. 2 , e105 (2006).
Eriksson, A. & Manica, A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc. Natl Acad. Sci. USA 109 , 13956–13960 (2012).
Burger, J., Kirchner, M., Bramanti, B., Haak, W. & Thomas, M. G. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc. Natl Acad. Sci. USA 104 , 3736–3741 (2007).
Malmström, H. et al. in Migration in Prehistory: DNA and Stable Isotope Analysis of Swedish Skeletal Material (ed. Linderholm, A.) (Stockholm University, 2008).
Malmström, H. et al. High frequency of lactose intolerance in a prehistoric hunter-gatherer population in northern Europe. BMC Evol. Biol. 10 , 89 (2010).
Lacan, M. et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc. Natl Acad. Sci. USA 108 , 9788–9791 (2011).
Plantinga, T. S. et al. Low prevalence of lactase persistence in Neolithic South-West Europe. Eur. J. Hum. Genet. 20 , 778–782 (2012).
Bollback, J. P., York, T. L. & Nielsen, R. Estimation of 2 N es from temporal allele frequency data. Genetics 179 , 497–502 (2008).
Malaspinas, A.-S., Malaspinas, O., Evans, S. N. & Slatkin, M. Estimating allele age and selection coefficient from time-serial data. Genetics 192 , 599–607 (2012).
Mathieson, I. & McVean, G. Estimating selection coefficients in spatially structured populations from time series data of allele frequencies. Genetics 193 , 973–984 (2013).
Steinrücken, M., Bhaskar, A. & Song, Y. S. A novel spectral method for inferring general diploid selection from time series genetic data. Ann. Appl. Statist. 8 , 2203–2222 (2014).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522 , 207–211 (2015).
Yang, Z. & Rannala, B. Bayesian species delimitation using multilocus sequence data. Proc. Natl Acad. Sci. USA 107 , 9264–9269 (2010).
Download references
Acknowledgements
The author would like to acknowledge Kelley Harris for helpful discussions regarding SMC-based approaches.
Author information
Authors and affiliations.
Department of Genome Sciences, University of Washington, 3720 15th Avenue NE, box 355065, Seattle, 98195–5065, Washington, USA
Joshua G. Schraiber & Joshua M. Akey
You can also search for this author in PubMed Google Scholar
Corresponding authors
Correspondence to Joshua G. Schraiber or Joshua M. Akey .
Ethics declarations
Competing interests.
J.M.A. is a paid consultant of Glenview Capital. J.G.S. declares no competing interests.
Supplementary information
Supplementary information s1 (box).
Issues in inferring absolute dates (PDF 141 kb)
Supplementary information S2 (box)
Lambda coalescents (PDF 139 kb)
PowerPoint slides
Powerpoint slide for fig. 1, powerpoint slide for fig. 2, powerpoint slide for fig. 3, powerpoint slide for table 1.
(EDA). The initial stages of 'digging into' a data set, usually by plotting low-dimensional summaries of the data.
The probabilities of the data given various models and their parameters, thought of as functions of those parameters. The parameter values that maximize the probability of the data in each model are called maximum likelihood estimates.
Vectors that, when multiplied by a given matrix, still point in the same direction.
An n×n matrix describing the covariance between each pair in a sample of size n .
A group of individuals among whom random mating occurs.
(LD). Nonrandom association between alleles at physically distinct genomic loci. Over time, this will be broken down by recombination.
The times in the past when genomic regions shared a common genetic ancestor.
Genetic differentiation between individuals induced by geographic separation. Individuals that are closer geographically will be closer genetically.
By adding more parameters to a model, it will begin to model the noise in the observed data, rather than the true underlying mechanism of data generation. Overfit models will generalize poorly to new data sets.
The error in predicting the structure of a held-out portion of the data, when a model is trained on a subset of the whole data set. Minimizing cross-validation error is an effective way to choose parameters and hyperparameters.
Graph structures representing the genealogical history of a sample with a recombining genome. In addition to coalescence events (which bring two lineages together and therefore reduce the number of lineages in the graph), recombination events cause splits to occur, which increases the number of lineages in the graph.
(HMM). A statistical model in which a set of underlying hidden states are assumed to follow Markov chain dynamics and induce a set of observed states.
A large number of individuals, related to samples of interest, for which some quality is known (for example, allelic phase).
The size that a theoretical population evolving under a Wright–Fisher model would need to be in order to match aspects of the observed genetic data.
A stochastic process in which new events occur at a constant rate per unit of time. Often used to model mutation.
(IBD). Whether a genomic region has descended from an ancestor unchanged. A genomic region in two (or more) individuals is identical by descent if it is inherited from a common ancestor without being broken up by recombination. Some authors require IBD segments to also be identical by state, that is, to also have no mutations in the region.
(IBS). Whether a genomic region has the same sequence as the corresponding region in another individual. A genomic region in two (or more) individuals is identical by state if it contains no mutations that distinguish the two individuals. Note that a region of IBS is not necessarily also identical by descent.
A modification to the sequential Markov coalescent (SMC) that allows for hidden recombination events that do not change the local genealogy.
Alleles that are sampled from a population given that a set of reference alleles is already in hand.
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Schraiber, J., Akey, J. Methods and models for unravelling human evolutionary history. Nat Rev Genet 16 , 727–740 (2015). https://doi.org/10.1038/nrg4005
Download citation
Published : 10 November 2015
Issue Date : December 2015
DOI : https://doi.org/10.1038/nrg4005
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Population genomics unravels the holocene history of bread wheat and its relatives.
Nature Plants (2023)
Olfactory marker protein contains a leucine-rich domain in the Ω-loop important for nuclear export
- Noriyuki Nakashima
- Akiko Nakashima
- Makoto Takano
Molecular Brain (2022)
Genome-wide association studies of yield-related traits in high-latitude japonica rice
- Guomin Zhang
- Rongsheng Wang
BMC Genomic Data (2021)
Admixture-enabled selection for rapid adaptive evolution in the Americas
- Emily T. Norris
- Lavanya Rishishwar
- I. King Jordan
Genome Biology (2020)
Whole-genome sequencing of 128 camels across Asia reveals origin and migration of domestic Bactrian camels
Communications Biology (2020)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- ORIGINAL SCIENTIFIC ARTICLE
- Open access
- Published: 29 July 2010
Human Origins Studies: A Historical Perspective
- Tom Gundling 1
Evolution: Education and Outreach volume 3 , pages 314–321 ( 2010 ) Cite this article
33k Accesses
1 Citations
2 Altmetric
Metrics details
Research into the deep history of the human species is a relatively young science which can be divided into two broad periods. The first spans the century between the publication of Darwin’s Origin and the end of World War II. This period is characterized by the recovery of the first non-modern human fossils and subsequent attempts at reconstructing family trees as visual representations of the transition from ape to human. The second period, from 1945 to the present, is marked by a dramatic upsurge in the quantity of research, with a concomitant increase in specialization. During this time, emphasis shifted from classification of fossil humans to paleoecology in which hominids were seen as parts of complex evolving ecosystems. This shift is in no small part due to the incorporation of neo-Darwinian synthetic theory. Finally, technological innovation and changes in social context are considered as influences on human origins studies.
Introduction
Considering the grand sweep of history, the realization that human beings gradually evolved from some non-human ancestor represents a very recent insight. Even so, the goal of this one brief essay cannot be to provide an in-depth description and analysis of every significant development within the field of paleoanthropology, but rather to identify broad patterns and highlight a collection of “events” that are most germane in shaping current understanding of our evolutionary origin. These events naturally include the accretion of fossil material, the raw data which is the direct, if mute, testimony of the past. These fossil discoveries are situated among technological breakthroughs, theoretical shifts, and changes in the sociocultural context in which human origins studies were conducted. It is only through such a contextualized historical approach that we can truly grasp our current understanding of human origins. Foibles of the past remind us to be critical in assessing newly produced knowledge, yet simultaneously we can genuinely appreciate the enormous strides that have been made.
In addition to selective coverage, a second caveat is that this review will focus on research by scientists writing in English. Non-modern hominids Footnote 1 are a cosmopolitan bunch, having been discovered throughout Africa, Asia, and Europe, and there is a significant literature in other languages. In an effort to ameliorate both of these shortcomings, numerous secondary references are included in the bibliography, providing more in-depth information on specific topics. For example, some texts approach the history of paleoanthropology by detailing a single time period (Bowler 1986 ), early human species (Walker and Shipman 1996 ), or researcher (Morell 1995 ), and there are quite a few that consider the subject more comprehensively (Leakey and Goodall 1969 ; Reader 1988 ; Lewin 1997 ; Tattersall 2008 ). In addition, there are a handful of encyclopedia format tomes (Jones et al. 1994 ; Spencer 1997 ; Delson et al. 2000 ), textbooks (Conroy 2005 ; Cela-Conde and Ayala 2007 ; Klein 2009 ), and “coffee table” popular volumes (Stringer and Andrews 2005 ; Johanson and Edgar 2006 ) that in part address the history of human origins studies. Moreover, these texts contain abundant references to the primary literature if that level of scrutiny is desired.
In seeking to provide a useful heuristic framework for the purposes of this particular essay, human origins studies can be broken down into two very broad periods. The first is roughly the century between 1850 and 1950 when research, often conducted by individuals with training outside of anthropology, focused on taxonomy and phylogeny. In other words, although scientists were cognizant that climate change (e.g., northern hemisphere glaciations) would have directly impacted the evolution of early humans, they were mainly interested in collecting “missing links,” naming them, and creating family trees. The second period, from 1950 to the present, is characterized by the relatively rapid development of paleoanthropology as it is currently practiced. Here the emphasis is only partly on the hominids themselves, with ecological context being of equal importance.
The Emergence of Human Origins Studies
This review begins with two mid-nineteenth century developments which are often conflated, but were initially distinct. The first is the acceptance of a temporal association of human material culture (stone tools), with extinct Ice Age mammals (Van Riper 1993 ; Sommer 2007 ). This was significant in that it opened up a considerable prehistory for the human species, well beyond estimates derived from literal scriptural interpretation. However, while acknowledging a lengthy antiquity for the human species, there was, at the time, no reason to suspect that the makers of the stone tools were not fully modern humans in a biological sense. The second major development was the publication of Darwin’s On the Origin of Species in 1859 (Darwin 1859 ). Darwin is rightfully credited with being the most influential, although by no means the first individual to broach the subject of descent with modification, or transmutation theory, as he put it (for an overview of pre-evolutionary ideas related to human origins, see Greene 1959 , Bowler 2003 ). Darwin’s central thesis was that all living species shared a common ancestry, with “endless forms most beautiful” having diverged via natural selection, and although he only briefly mentioned his own species the inference was clear. These two events dovetailed into the now quotidian, but then controversial, notion that humans had evolved over a vast expanse of time (Grayson 1983 ).
While Darwin was initially reticent to discuss human evolution in any detail, his colleague Thomas H. Huxley harbored no such reluctance when he published Man’s Place in Nature: Essays in 1863 (Huxley 1900 ). Darwin freely admitted that the veracity of his audacious proposal would have to withstand paleontological scrutiny and that his theory would collapse in the absence of transitional fossil forms. Huxley’s advantage, beyond his more outspoken personality, was that he actually had a fossil human to describe. The first Neandertal recognized by science was discovered in 1856; however, its description only appeared in English three years later, just as Darwin was going to press (Trinkaus and Shipman 1993 ). Huxley provided a detailed description of the eponymous cranium coupled with carefully composed line drawings (Huxley 1900 ). However, while the importance of the Neandertals in providing empirical evidence documenting an ancient and morphologically distinct human form cannot be discounted, these people hardly bridged the gap separating humans and the great apes. Although a few dissenting voices denied the close evolutionary relationship among humans and the “man-like” apes, and consequently an ape phase of human ancestry, most scientists accepted the overwhelming morphological and embryological evidence in support of just such a relationship. This acceptance was in no small part due to Huxley’s meticulous comparison of gorilla and human anatomy in which he concluded that the gorilla and its close relation, the chimpanzee, represented the nearest approach to humanity in nature.
If Neandertals were more or less human, then more distant, primitive “missing links” remained to be discovered. Just such fossils were recovered on the island of Java in the 1890s by Dutch physician Eugene Dubois, who had traveled to Indonesia as part of the army but with the express purpose of finding the remains of primitive humans (Shipman 2001 ). Java Man consisted of a skull cap, a femur, and a few isolated teeth which taken in combination suggested an early human with a much smaller cranial capacity relative to Neandertals or Homo sapiens (roughly 1,000 vs. 1,500 cubic centimeters), although the femur appeared modern. Dubois did not receive the universal accolades and acceptance he coveted, but his fossils bolstered the conventional wisdom at the time that humans first evolved somewhere in Asia.
During the early twentieth century, the early hominid fossil record grew significantly, if not exponentially, and evolution was widely accepted in scientific circles even while large segments of the lay public remained skeptical. Certainly, there were disagreements over whether natural selection was a sufficient evolutionary mechanism in itself (Bowler 1983 ), but the basic premise of biological change through time was affirmed. The recovery of additional Neandertal remains in Europe refuted lingering claims of pathology regarding the original Neander Valley specimen and solidified the interpretation that the latter was representative of a population of archaic humans occupying Ice Age Europe. Some Neandertal remains were interpreted as not only indicating intentional internment but also associated funerary ritual. The European fossil record was extended significantly with the recovery of a robust lower jaw from Mauer, Germany discovered in 1907.
In 1912 in England, heretofore devoid of non-modern hominid remains despite the prominence of several British scholars in human origins studies, the announcement of hominid fossils from Piltdown was warmly received locally, if with some incredulity abroad. Piltdown was significant since it reified the “brain first” hypothesis, in which primitive humans evolved a large brain before other key human traits evolved. Although a favorite of intelligent design creationism advocates, Piltdown is actually a beautiful example of the scientific method at work, whereby new evidence eventually calls into question prior interpretation, and in this case recognition of intentional fraud (Spencer 1990 ). It was, after all, a new relative dating method measuring the fluorine content of fossils that in 1953 exposed the non-contemporaneity of the jaw and skull. In any case, in the first decades of the twentieth century, a fairly simple human family tree was beginning to emerge (see McCown and Kennedy 1972 and especially Delisle 2007 for exceptions). Relatively small-brained Pithecanthropus led to the more capacious Neandertals and Piltdown, who in turn evolved into modern H. sapiens . Yet the truly ape-like human ancestors remained elusive.
Africa as the Cradle of Humanity
In 1921 a skull bearing superficial resemblance to European Neandertals was recovered as part of mining operations at a place called Broken Hill in Northern Rhodesia (now Zambia). Rhodesian Man marks the recovery of the first in a very long line of non-modern hominids from the African continent. A mere four years later, University of Witwatersrand anatomist Raymond Dart, Australian by birth and having been trained in England, published a brief paper describing the fossil skull of a juvenile “ape” discovered in a limestone quarry near Taung, South Africa. Dart identified certain features of the face, the teeth, the cranium, and the brain of Australopithecus africanus that foreshadowed those of H. sapiens and made the startling claim that what was essentially a bipedal ape signaled the beginning of the human lineage separate from the African great apes.
Initially, with only the one individual, and a juvenile at that, Dart found little support. His most ardent advocate, Scottish physician and paleontologist Robert Broom discovered additional fragmentary remains of the australopithecines, as they were then called, in other South African caves in the 1930s, but these were initially insufficient to sway opinion (Dart 1959 ). This was perhaps due to the near-simultaneous discovery of significant hominid remains from Zhoukoutien (Dragon Bone Hill) in China which quickly eclipsed whatever controversy the diminutive skull from Taung elicited, and despite Broom’s ongoing efforts. As was the case with Java Man, the more complete Chinese fossils fulfilled the expectations of many scientists who anticipated that earliest human ancestors evolved to the East. Comparative analysis of the Javanese and Chinese fossils revealed a great deal of similarity, and all of the fossils were ultimately subsumed in the species Homo erectus .
The Neo-Darwinian Synthesis and the New Physical Anthropology
For several disparate reasons, the decades following the end of World War II (WWII) rather quickly led to a science of paleoanthropology that is recognizably modern. One significant factor relevant in the U.S. if not everywhere, was the dramatic upsurge in enrollment at colleges and universities. The G.I. Bill and subsequent effects of Civil Rights legislation that greatly increased access to higher education meant that millions more students went to college and hence the expansion of existing campuses and programs and in some cases the appearance of entirely new colleges and universities Footnote 2 . As a result, greater numbers of faculty were required who could teach courses and supervise research in diverse academic programs, which in turn led to an attendant rise in the numbers of graduate students themselves who went on to secure positions at institutions of higher learning. Consequently, many disciplines experienced significant increases in research activity, including physical anthropology, and it is worth noting that this was the first generation of researchers whose formal training was in physical anthropology, not in some allied field such as anatomy or medicine. The dramatic rise in practitioners not only increased the knowledge base in terms of simple quantity, but specialization within the field also began to emerge.
A second crucial development that transformed human evolutionary studies was theoretical in nature. Changing ideas regarding the process of evolution had been fermenting and roiling in biology circles for several decades before they infiltrated the study of human origins. In essence, a consensus was reached among biologists ( sensu lato ) that Darwinian natural selection acting on variation arising from random mutation was a sufficient mechanism to explain evolutionary change. For anthropologists, although questions of taxonomy and phylogeny remained important, the intellectual fallout of the so-called neo-Darwinian synthesis led to the “New Physical Anthropology” in which early hominid fossils, rather than representative of some platonic archetype, were interpreted as unique members of variable populations. Focusing on evolution as a process effecting change in populations over time, in contrast to the comparatively myopic sorting of the resulting pattern , arguably represents the most significant theoretical shift in thinking about evolution since Darwin.
Given the comparative de-emphasis on iconic types, the bloated alpha taxonomy of the past was reduced to a mere handful of hominid species displaying previously under-appreciated within species variability. This great reduction in hominid names and consequent simplification of hominid family trees has led some modern scholars to lament what they see as a return to the bad old days of teleology and orthogenesis. Yet there can be little doubt that the “splitting” taxonomic philosophy of the past where almost every new specimen received a new species or quite frequently a new genus name was in dire need of revision.
Just as species types came under scrutiny, so did the concept of evolutionary grades which had up to this point made clear distinctions between the categories of ape and human. While this may have provided some welcome taxonomic clarity, it was artificial in that it ignored the evolutionary reality that at some point members of the human lineage were very ape-like. This realization, obvious in retrospect, led to the widespread acceptance of the South African australopithecines as human ancestors, and the important corollary that bipedalism preceded other distinctive human attributes (Gundling 2005 ).
In addition to increased research activity and theoretical shifts, by the early 1960s technological innovations for the first time permitted the creation of a reliable absolute timescale of human evolution. Comparative protein analysis demonstrated that the African apes were most similar genetically to H. sapiens , inferring their recent common ancestry to the exclusion of other apes and monkeys. Molecular clocks based upon mutation rates and calibrated by the fossil record suggested that this common ancestor lived as little as a few million years ago, although recent estimates put this ancestor at seven to five million years ago. Consequently, known early and middle Miocene ape species became suspect as purported human ancestors, since they preceded the split between the hominid and great ape lineages. Most notably this eventually led to the downfall of Ramapithecus , a Miocene ape genus once widely hailed as a very ancient and very primitive hominid Lewin ( 1997 ).
While molecular studies of living species effectively imposed a theoretical maximum on the age of the hominid lineage, the temporal framework of human origins was further clarified with the introduction of the new potassium argon (K-Ar) method of absolute dating. Louis and Mary Leakey had been scouring the fossil-bearing sediments in and around eastern Africa’s Great Rift Valley for decades when Mary discovered the skull of a robust australopithecine at Olduvai Gorge in 1959. Significantly, Zinjanthropus , the genus coined for the new skull, was discovered within sediments near the base of the Pleistocene Epoch. Volcanic minerals from associated strata were dated to approximately 1.75 million years ago using the K–Ar method, nearly double the age estimated from using other more crude means. This greatly expanded time range certainly bolstered claims for the australopithecines as human ancestors rather than extinct collateral cousins to the “true” human lineage, yet to be discovered.
As an aside, Louis Leakey’s interest in understanding the human past was not limited to the collection of fossils. Sherwood Washburn, a main architect of the new physical anthropology, along with Irven DeVore, conducted pioneering studies of savanna baboons, large-bodied, terrestrial, and highly social primates that served as living proxies for modeling early hominid behavioral ecology (Washburn and DeVore 1961 ). Leakey, on the other hand, took a more phylogenetically based approach and hired scholars to conduct research into the behavior of the great apes as a potential new data source informing hypotheses of early hominid behavior. Jane Goodall was the first, studying chimpanzee behavior at Gombe in Tanzania, then came Dianne Fossey who undertook a longitudinal study of mountain gorillas in Rwanda, and finally Birute Galdikas traveled to Indonesia to conduct field studies of the orangutan (see Kinzey 1987 and De Waal 2001 for more recent primate studies that explicitly address questions of human behavioral evolution).
The emergence of paleoanthropology as a truly multidisciplinary endeavor, concerned with a more holistic picture of our evolutionary past, was a logical extension of the post-WWII new physical anthropology which eschewed simple classification and promoted variable populations as the unit of study. Naturally, these hominid populations did not exist in a vacuum but were components of complex, evolving ecosystems. Hence, field work began to emphasize the collection of greater contextual data in an effort to reconstruct biological and physical environments in which these human ancestors existed and evolved. One of the first field projects to adopt this new approach was an international expedition centered around the Omo River Valley in southern Ethiopia, beginning in 1967. Remarkably, of the 50 papers collected in the resulting volume, only five primarily focus on the hominid remains themselves (Coppens et al. 1976 ).
Early Human Diet and Subsistence
One major aspect of early hominid ecology that occupied researchers engaged in such multidisciplinary efforts was subsistence, which has understandably been of great interest to paleoanthropologists, particularly after 1950 as scientists endeavored to contextualize the fossil remains of distant ancestors. What early humans ate, how food was acquired and processed, even how it was distributed among members of a social group, became viable questions. Throughout the 1950s and 1960s it was widely assumed that the social, cognitive, and technological skills associated with big-game hunting drove the evolution of the human species; in fact the allure of “Man the Hunter” is longstanding in Western thought (Cartmill 1993 ). Raymond Dart, as part of his second foray into human origins studies, proposed that Australopithecus had already developed a hunting strategy facilitated by a technology comprised of durable animal parts that he referred to as the osteodontokeratic (bone, tooth, horn) culture. This concept was enthusiastically embraced by writer Robert Ardrey, who published a series of four popular novels documenting the success of these “killer apes” in the context of a changing environment (e.g., Ardrey 1976 ). Research scientists were only slightly less enthusiastic in championing such ideas (Lee and DeVore 1968 ) which remain popular, if more nuanced today (Wrangham and Peterson 1996 ).
Mirroring changes in the broader society, by the early 1970s some anthropologists challenged the “Man the Hunter” hypothesis and developed an alternative that focused on the central role of women in child rearing and gathering of food resources (Dahlberg 1981 ). These studies used ethnographic data from extant food-foraging societies, the rarity of which injected a sense of urgency on the part of anthropologists. Not long after the “Women the Gatherer” model appeared as a second wave feminist rejoinder to the previously unquestioned authority of “Man the Hunter,” another group of researchers also began to question the big-game hunting scenario. Archeologists, geologists, and paleontologists began working on “site formation processes” to get a better understanding of how assemblages of fragmented animal bones and stone tools came to be commingled. Over the next few decades, often with recourse to modern ecosystems as analogs, one of the main conclusions drawn from the new science called taphonomy (=laws of burial) was the potential importance of scavenging. The association of “bones and stones” was no longer assumed to be the signature of hominid big-game hunting but instead interpreted as meals containing essential fat and protein scavenged by early humans. Perhaps even more disconcerting, some sites were reinterpreted as the remains of carnivore kills occasionally including early humans themselves (Brain 1981 ; Hart and Sussman 2008 ).
Here’s Lucy
If Mary and Louis Leakey’s discoveries at Olduvai put the Great Rift Valley on the map, during the 1970s eastern Africa was validated as the center of early hominid studies. The Leakey’s son Richard established himself on the east side of Lake Turkana in northern Kenya, where his expeditions uncovered a prolific cache of early hominid fossils, some of which corroborated the occasionally controversial claims made by his parents a decade earlier. Sediments around the lake yielded hominid fossils of robust australopithecines, early members of genus Homo , and an early African variant of Asian H. erectus , these days referred to as Homo ergaster (Leakey and Lewin 1978 ). The latter includes a mostly complete skeleton, KNM-WT15000, which has become iconic for the species (Walker and Shipman 1996 ).
Arguably the most significant fossil discovery of the 1970s was another partial skeleton, AL-288, from Hadar, Ethiopia, better known as Lucy (Johanson and Edey 1981 ). Here was a single individual represented by numerous skeletal elements, and although her morphology was generally similar to the “gracile” australopithecines of South Africa, she was even more primitive in some respects. Consequently her discoverers coined a new species name, Australopithecus afarensis that included not only the Hadar specimens but fossils collected by Mary Leakey’s expedition at Laetoli in Tanzania. The latter is renowned for its famous footprint trail preserved in solidified volcanic ash, imparting convincing evidence for bipedalism at 3.6 million years ago. Hadar is also replete with datable volcanic sediments, and Lucy’s status as the most primitive hominid was reinforced by firm radiometric dates which placed the fossils at greater than 3.0 million years ago, at the time astonishingly ancient.
One other significant event from the 1970s bears mentioning. Although the American Journal of Physical Anthropology was first published in 1918, it is perhaps surprising that a journal explicitly dedicated to the study of human evolution did not appear in the U.S. until 1972. Since then the Journal of Human Evolution has been the premier academic forum for publications related to human evolution, and in 1992, the Paleoanthropology Society was established, which organizes its own conference and publishes an online journal.
Modern Human Origins
The question of modern human origins has been debated for centuries, long predating paleoanthropology as a scientific discipline. One of the central issues, which became particularly evident as Renaissance and Enlightenment Europeans began to travel the globe on a regular basis, was how to explain the physical diversity of human populations. Two broad perspectives emerged, one which viewed all people as having a single origin and another which believed that supposedly distinct races had separate origins. The pre-Darwinian debates between so-called monogenists and polygenists were recast with the advent of an evolutionary paradigm in the mid-nineteenth century. Within this new theoretical context, monogenists believed that all living humans evolved from a common ancestor that was already H. sapiens , while the polygenists believed that the races had deeper roots and had descended from different non-modern ancestors (e.g., H. erectus or in a few instances different ape species). A major step towards resolving this debate came in 1987 with an analysis of living human mitochondrial DNA diversity which concluded that H. sapiens had a recent African origin. The discovery of essentially modern human fossils at the 160,000-year-old site of Herto, Ehtiopia, provides paleontological support for a recent African origin, and many subsequent genetic studies have supported this basic conclusion. However, the possibility of some gene flow between migrating early modern humans and local archaic populations remains plausible (compare Stringer and McKie 1996 and Wolpoff and Caspari 1998 , also see Relethford 2003 for a geneticist’s perspective).
Conclusion: Twenty-First Century Paleoanthropology
New fossil discoveries, technological innovations, theoretical advances, and social transformations will continue to inform knowledge of our deep past. Recovery of hominid fossils, some from previously unknown time periods and geographic locations, continues at a brisk rate. Many of the most significant recent discoveries are beginning to fill in the crucial African late Miocene time period during which our lineage ramified from that leading to the chimpanzee (Gibbons 2006 ). Of particular note, one of these fossils was discovered in Chad, quite a distance from established sites in the Great Rift Valley, challenging the long standing hypothesis that hominids evolved in the savanna grasslands of eastern Africa while the African ape ancestors remained sequestered in their tropical rainforest refugium. Moreover, botanical, faunal, and geological evidence associated with very early fossil hominids in Ethiopia and Kenya intimate a forested environment, a discovery that clearly constrains hypotheses explaining the success of the bipedal adaptation.
Other significant fossil discoveries from the early Pleistocene site of Dmanisi in the Republic of Georgia have energized discussion of the initial expansion of early humans beyond the tropics of Africa (Wong 2006 ). Not only are these fossils considerably older than prior known Eurasian specimens, but they are morphologically primitive, especially in terms of stature and cranial capacity, and are associated with very simple (“mode 1”) lithic technology. These early migrants hardly manifest the tall striding bipeds equipped with comparatively advanced Acheulian bifacial tools so often depicted in earlier “out of Africa” scenarios Footnote 3 , which are at least in part based on the iconic WT15000 skeleton mentioned earlier.
Perhaps the most surprising discovery of the last decade is the diminutive 18,000-year-old skeleton from the Indonesian island of Flores, which has sparked a spirited, occasionally acrimonious debate between those advocates of a replacement model of modern human origins and those inclined towards regional continuity (Morwood and van Oosterzee 2007 ). The former, comprised of the team who made the discovery and their allies, interpret the remains as those of a surprisingly primitive hominid akin to early Homo , and perhaps the first documented example of the effects of island dwarfing on an early human population. Other scholars believe the remains to be those of a pathological modern human, whose illness resulted in a cascade of skeletal and dental anomalies. Ongoing research on Flores and other nearby locations will undoubtedly resolve this debate.
New discoveries are not limited to the paleontological record but also include behavioral information gleaned from archaeology. Symbolic expression in the form of language, art (including music), and religion is undoubtedly one of the most distinctive human traits. Evidence for such behavior has proved elusive beyond the seeming cultural explosion perceived in the Upper Paleolithic of Europe beginning around 35,000 years before present. However, archeological evidence for at least some of these behaviors has recently been coaxed out of several sites in sub-Saharan Africa. Advanced utilitarian objects such as blades and harpoons have been recovered well back into the Middle Stone Age and use of ochre and shells for body adornment has been found at sites approaching 100 kiloannum (Balter 2009 ).
Recent advances also include a plethora of technological innovations that have allowed anthropologists to hone traditional inquiries in the areas of dating (e.g., single crystal, laser fusion, argon–argon dating), systematic analysis (e.g., geometric morphometrics), and paleoenvironmental reconstruction (e.g., stable isotope analysis). The badly distorted remains of the spectacular 4.4 megaanum skeleton of Ardipithecus ramidus from Aramis, Ethiopia was restored in part using digital imaging technology (Gibbons 2009 ). Additionally, new technology is facilitating, perhaps even driving, novel questions such as those related to the emergence of the unique human life history pattern.
While fossils provide real-time evidence for human evolution, signals from our ancient past are also encoded into our modern DNA. The groundbreaking work of the 1960s effectively demonstrated our close affinity with the African great apes, and today’s genomic analyses comparing humans and chimpanzees are beginning to reveal differences in much finer detail than heretofore possible. Already several areas within the human genome have been identified as having undergone intense selection; these regions may be related to the evolution of the especially dexterous human thumb, reduction of muscles of mastication in the wake of the ability to cook food, the greatly enlarged neo-cortex, and our ability for spoken language.
In addition to modern DNA analyses, ancient DNA analysis has informed the “Neandertal problem” providing preliminary evidence in support of the replacement hypothesis, at least in Europe, whereby modern humans arriving there equipped with Upper Paleolithic technology drove the indigenous Neandertals to extinction. Even more recent genomic analyses, however, suggest that a small but detectable degree of interbreeding occurred when expanding modern human populations emerging from the African tropics encountered Neandertal populations in the Middle East around 120,000 years before present (Gibbons 2010 ).
In conclusion, our understanding of human origins, like all scientific knowledge, is the result of an ongoing, iterative process. Over the last few decades, the accelerating pace of fossil discoveries and the incorporation of innovative technologies have corroborated and enhanced much of what we already suspected to be true, although there have been a few surprises. No doubt this pattern will continue into the foreseeable future as we slowly, yet inexorably, piece together the circumstances by which our lineage became human.
Hominidae (=hominid) is the biological group (clade) to which humans and their extinct ancestors belong. For many current scholars, this group is distinguished at some lower taxonomic level, usually the tribe Hominini (=hominin). In this study, I maintain the traditional use of Hominidae simply to be consistent with the historical literature. For the same reason, I use the subfamily designation Australopithecinae (=australopithecine) for all of the African “bipedal apes.” This group is certainly paraphyletic, to use the modern jargon, and as a result an increasing number of scholars prefer to use the less formal term australopith to lump together the various African species.
According to the U.S. Census Bureau, there were about 2.2 million students enrolled in U.S. colleges in 1950. That number doubled by 1963 to just less than 4.4 million and doubled again to over 9 million by 1972.
I usually avoid this term despite its heavy usage within the scientific community. I believe that the “Out of Africa” trope perpetuates an anti-Africa bias which seems to suggest that early humans, on several occasions, left Africa wholesale as if there was something inherently undesirable about the place. I suppose that “hominid extra-tropical range expansion” doesn’t have the same ring, but it is more accurate.
Ardrey R. The hunting hypothesis. New York: Atheneum; 1976.
Google Scholar
Balter M. On the origin of art and symbolism. Science. 2009;323:709–11.
Article CAS PubMed Google Scholar
Bowler PJ. The eclipse of darwinism. Anti-darwinian evolution theories in the decades around 1900. Baltimore: The Johns Hopkins University Press; 1983.
Bowler PJ. Theories of human evolution: a century of debate 1844–1944. Baltimore: The Johns Hopkins University Press; 1986.
Bowler PJ. Evolution: the history of an idea. 3rd ed. Berkeley: University of California Press; 2003.
Brain CK. The hunters or the hunted? An introduction to African cave taphonomy. Chicago: University of Chicago Press; 1981.
Cartmill M. A view to a death in the morning: hunting and nature through history. Cambridge: Harvard University Press; 1993.
Cela-Conde CJ, Ayala FJ. Human evolution: trails from the past. Oxford: Oxford University Press; 2007.
Conroy GC. Reconstructing human origins. 2nd ed. New York: W.W. Norton; 2005.
Coppens Y, Howell FC, Isaac GL, Leakey REF. Earliest Man and environments in the lake rudolf basin: stratigraphy, paleoecology and evolution. University of Chicago Press; 1976.
Dahlberg F, editor. Woman the gatherer. New Haven: Yale University Press; 1981.
Dart RA. Adventures with the missing link. Britain: Hamish Hamilton, Ltd.; 1959.
Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859.
Book Google Scholar
De Waal FB, editor. Tree of origin: what primate behavior can tell us about human social evolution. Cambridge: Harvard University Press; 2001.
Delisle RG. Debating humankind’s place in nature 1860–2000: the nature of paleoanthropology. Upper Saddle River: Pearson Education, Inc.; 2007.
Delson E, Tattersall I, Van Couvering JA, Brooks AS. Encyclopedia of human evolution and prehistory. 2nd ed. New York: Garland Publishing, Inc.; 2000.
Gibbons A. The first human: the race to discover our earliest ancestors. New York: Doubleday; 2006.
Gibbons A. Breakthrough of the year: Ardipithecus ramidus . Science. 2009;326:1598–9.
Gibbons A. Close encounters of the prehistoric kind. Science. 2010;328:680–4.
Grayson DK. The establishment of human antiquity. New York: Academic; 1983.
Greene JC. The death of Adam: evolution and its impact on western thought. Ames: Iowa State University Press; 1959.
Gundling T. First in line: tracing our ape ancestry. New Haven: Yale University Press; 2005.
Hart D, Sussman RW. Man the hunted: primates, predators, and human evolution. Expandedth ed. Boulder: Westview Press; 2008.
Huxley TH. Man’s place in nature: essays. New York: D. Appleton & Co.; 1900 [1863]
Johanson D, Edey M. Lucy: the beginnings of humankind. New York: Simon and Schuster; 1981.
Johanson DJ, Edgar B. From Lucy to language. Revised, updated, and expanded. New York: Simon & Schuster; 2006.
Jones S, Martin R, Pilbeam D, editors. The cambridge encyclopedia of human evolution. Cambridge: Cambridge University Press; 1994.
Kinzey WG, editor. The evolution of human behavior: primate models. Albany: SUNY Press; 1987.
Klein RG. The human career: human biological and cultural origins. 3rd ed. Chicago: University of Chicago Press; 2009.
Leakey LSB, Goodall VM. Unveiling man’s origins. Cambridge: Schenkman Publ. Co.; 1969.
Leakey RE, Lewin R. People of the lake: mankind and its beginnings. New York: Doubleday; 1978.
Lee RB, DeVore I, editors. Man the hunter. Chicago: Aldine Publishing Co.; 1968.
Lewin RL. Bones of contention: controversies in the search for human origins. 2nd ed. Chicago: University of Chicago Press; 1997.
McCown TD, Kennedy KAR, editors. Climbing man’s family tree: a collection of major writings on human phylogeny, 1699 to 1971. Englewood Cliffs: Prentice Hall; 1972.
Morell V. Ancestral passions: the Leakey family and the quest for humankind’s beginnings. New York: Simon & Schuster; 1995.
Morwood MJ, van Oosterzee P. A new human: the startling discovery and the strange story of the “Hobbits” of Flores, Indonesia. New York: Harper Collins; 2007.
Reader J. Missing Links: the hunt for earliest man. 2nd ed. London: Pelican Books; 1988.
Relethford JH. Reflections of our past: how human history is revealed in our genes. Boulder: Westview Press; 2003.
Shipman P. The man who found the missing link: Eugene Dubois and his lifelong quest to prove Darwin right. New York: Simon and Schuster; 2001.
Sommer M. Bones and ochre: the curious afterlife of the red lady of paviland. Cambridge: Harvard University Press; 2007.
Spencer F. Piltdown: a scientific forgery. London: British Museum (Natural History); 1990.
Spencer F, editor. History of physical anthropology: an encyclopedia. New York: Garland Publishing, Inc.; 1997.
Stringer C, Andrews P. The complete world of human evolution. London: Thames and Hudson; 2005.
Stringer C, McKie R. African exodus: the origins of modern humanity. New York: Henry Holt; 1996.
Tattersall I. The fossil trail: how we know what we think we know about human evolution. 2nd ed. Oxford: Oxford University Press; 2008.
Trinkaus E, Shipman P. The Neandertals: changing the image of mankind. New York: Alfred A. Knopf; 1993.
Van Riper AB. Men among mammoths: victorian science and the discovery of human prehistory. Chicago: Chicago University Press; 1993.
Walker A, Shipman P. The wisdom of the bones: in search of human origins. New York: Alfred A. Knopf; 1996.
Washburn SL, DeVore I. The social life of baboons. Sci. Am. June; 1961
Wolpoff M, Caspari R. Race and human evolution. Boulder: Westview Press; 1998 [1997]
Wong K. Stranger in a new land. Sci Am. 2006;16(2):38–47.
Article Google Scholar
Wrangham R, Peterson D. Demonic males: apes and the origins of human violence. New York: Houghton Mifflin; 1996.
Download references
Author information
Authors and affiliations.
Department of Anthropology, William Paterson University, 300 Pompton Road, Wayne, NJ, 07470, USA
Tom Gundling
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Tom Gundling .
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Reprints and permissions
About this article
Cite this article.
Gundling, T. Human Origins Studies: A Historical Perspective. Evo Edu Outreach 3 , 314–321 (2010). https://doi.org/10.1007/s12052-010-0248-7
Download citation
Published : 29 July 2010
Issue Date : September 2010
DOI : https://doi.org/10.1007/s12052-010-0248-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Paleoanthropology
- History of science
- Human evolution
Evolution: Education and Outreach
ISSN: 1936-6434
- Submission enquiries: [email protected]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.8(6); 2018 Mar
How scientists perceive the evolutionary origin of human traits: Results of a survey study
Hanna tuomisto.
1 Department of Biology, University of Turku, Turku, Finland
Matleena Tuomisto
Jouni t. tuomisto.
2 National Institute for Health and Welfare, Kuopio, Finland
Associated Data
Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.s9r98
Various hypotheses have been proposed for why the traits distinguishing humans from other primates originally evolved, and any given trait may have been explained both as an adaptation to different environments and as a result of demands from social organization or sexual selection. To find out how popular the different explanations are among scientists, we carried out an online survey among authors of recent scientific papers in journals covering relevant fields of science (paleoanthropology, paleontology, ecology, evolution, human biology). Some of the hypotheses were clearly more popular among the 1,266 respondents than others, but none was universally accepted or rejected. Even the most popular of the hypotheses were assessed “very likely” by <50% of the respondents, but many traits had 1–3 hypotheses that were found at least moderately likely by >70% of the respondents. An ordination of the hypotheses identified two strong gradients. Along one gradient, the hypotheses were sorted by their popularity, measured by the average credibility score given by the respondents. The second gradient separated all hypotheses postulating adaptation to swimming or diving into their own group. The average credibility scores given for different subgroups of the hypotheses were not related to respondent's age or number of publications authored. However, (paleo)anthropologists were more critical of all hypotheses, and much more critical of the water‐related ones, than were respondents representing other fields of expertise. Although most respondents did not find the water‐related hypotheses likely, only a small minority found them unscientific. The most popular hypotheses were based on inherent drivers; that is, they assumed the evolution of a trait to have been triggered by the prior emergence of another human‐specific behavioral or morphological trait, but opinions differed as to which of the traits came first.
1. INTRODUCTION
Human evolution is a topic that interests not just researchers specialized in paleoanthropology, but also other scientists and the general public. A number of conflicting hypotheses have been put forward to explain why humans have become strikingly different from other primates. Most scientists in relevant fields (such as paleoanthropology, paleontology, ecology, evolution and human biology) have never published their views on the drivers of human evolution in general, nor on which of the proposed hypotheses on the origin of specific human traits they find most substantiated. No recent summary of the mainstream view among paleoanthropologists has been published either, so there is uncertainty as to whether scientists agree on the driving forces behind human evolution or not. The idea of carrying out a survey to find out emerged when one of us was teaching a university course on human evolution, happened to check what Wikipedia had to say on the subject, and noticed that some Talk pages (especially the one behind the article “Aquatic ape hypothesis”) contained definite but unreferenced claims about what the opinions of “all scientists” or “all paleoanthropologists” are.
Humans differ from all the other 400 primate species in many respects, some of the most striking ones being that they walk fully upright on their hind legs, have unusually big brains, and have an effectively naked rather than fur‐covered skin (Figure 1 ). Other features that among primates are uniquely human include descended larynx, articulated speech and the capacity to accumulate fat in a thick subcutaneous layer.

Male and female human figures from the plaque of the Pioneer 10 and 11 spacecrafts. The pictorial message was intended to describe the origin of the probe for potential extraterrestrial life. It shows several typically human traits, such as bipedalism, nakedness, arched nose, large head, and opposable thumbs. Source: NASA ; vectors by Mysid (Public domain), via Wikimedia Commons
A number of conflicting hypotheses have been proposed to explain why these and other traits originally evolved in the lineage leading to humans but in none of the lineages leading to other extant primates. One line of argumentation is based on the widely accepted idea that animal species adapt to their environment by natural selection: Traits that give the animal a higher probability of survival and reproduction become more common over time and traits related to lower survival and reproduction rates become less common. Adaptive traits are often morphological (like long legs that increase running speed and facilitate escaping from predators, or thick fur that protects from heat loss in cold weather), but they can also be behavioral (like building a nest or being nocturnal). The corollary of viewing traits of a species as adaptations to its environment is that traits are expected to change if the environment changes, because then also the adaptive pressures change. In particular, if sister species have very different traits in spite of close genetic relatedness, the adaptationist scenario suggests that the lineages experienced different environments during their evolutionary past.
It has indeed been proposed that the ancestors of humans came to live in a different kind of environment than the ancestors of chimpanzees and gorillas, and adapted by evolving a suite of novel traits. One of the early proposals along these lines, suggested already by Lamarck and Darwin, was that human ancestors descended from the trees and moved to the open savanna (Bender, Tobias, & Bender, 2012 ; Dart, 1925 ; Domínguez‐Rodrigo, 2014 ; Leakey & Lewin, 1977 ). Because terrestrial life in the dry savanna is very different from arboreal life in wet forests, this change in habitat would have shifted the prevailing selection pressures: Traits that were adaptive in the old environment could become maladaptive in the new one, and novel morphological traits could be favored if they gave a higher probability of survival and reproduction. The ancestors of the great apes stayed in the forest and, therefore, remained more similar to other primates.
The savanna scenario has lost some of its appeal since paleoenvironmental reconstructions started to show that the environmental setting has been more complex than was originally thought. Accordingly, more recent accounts describe the environment of early human ancestors as a mosaic of woodlands, savanna, and water bodies with considerable temporal fluctuations between climatically arid and wet periods (Bender et al., 2012 ; Domínguez‐Rodrigo, 2014 ; Kingston, 2007 ; Kovarovic & Andrews, 2007 ; Maslin & Christensen, 2007 ). Environmental variability itself has also been proposed to have selected for versatility of adaptations (Potts, 1998a , b ).
There have been different views on which aspects of terrestrial life would have required the morphological changes that the human lineage has experienced, so a large number of different explanations have been put forward for each trait. For example, the origin of the bipedal gait has been attributed to (among other things) gaining better visibility over the savanna grass (Ravey, 1978 ), reaching for food on low branches (Hunt, 1994 , 1996 ), collecting small food items from the ground (Jolly, 1970 ; Kingdon, 2003 ), exposing a smaller part of the body to the scorching sun (Wheeler, 1984 , 1991 ), allowing more energy‐efficient long‐distance travel (Carrier et al., 1984 ; Pontzer, Raichlen, & Sockol, 2009 ; Rodman & McHenry, 1980 ), and freeing the hands to carry food, tools, weapons, or babies (Bartholomew & Birdsell, 1953 ; Hewes, 1961 ; Lovejoy, 1981 ; Sutou, 2012 ; Washburn, 1960 ). It has also been proposed that bipedalism originated already in the trees for hand‐supported walking on small branches too weak for brachiation (Crompton, Sellers, & Thorpe, 2010 ; Thorpe, Holder, & Crompton, 2007 ).
Another adaptationist proposal is that the human ancestors moved from the trees to the waterside, and started to adapt to a partly aquatic way of life (Hardy, 1960 ; Morgan, 1982 ; Verhaegen, Puech, & Munro, 2002 ). This would have exposed them to similar selection pressures than semi‐aquatic mammals, rather than to selection pressures typically experienced by other primates. Under this scenario, bipedal gait would have emerged because it allowed wading to deeper water and made the body more streamlined when swimming and diving for food (Kuliukas, 2002 ; Morgan, 1990 ; Niemitz, 2010 ; Verhaegen et al., 2002 ).
Not all traits need to have originated to enhance survival, however, and critical voices have been raised against interpreting all uniquely human traits as adaptations driven by natural selection (Gee, 2013 ). Sexual selection is known to have produced spectacular new traits in various animals, typically ornaments whose sole purpose is to attract the attention of the opposite sex. These confer no survival advantage or may even be harmful to the bearer. At least human bipedalism, nakedness, and subcutaneous fat layer have been explained by this mechanism (Barber, 1995 ; Giles, 2011 ; Tanner, 1981 ). Especially in small populations, traits may even emerge due to chance fixation of random variation (Sutou, 2012 ).
For someone interested in the “why” of human evolution, it is currently hard to find a comprehensive account of the scientific state of the art. Journal articles typically address only one or a few hypotheses in isolation of the others and often their focus is more on “how” than on “why” a given trait originally emerged (e.g., Crompton et al., 2010 ; Cunnane & Crawford, 2014 ; Isler & Van Schaik, 2014 ; Stout & Chaminade, 2012 ; Watson, Payne, Chamberlain, Jones, & Sellers, 2008 ; Wells, 2006 ). Only proponents of the aquatic/waterside hypotheses (collectively known as the aquatic ape hypothesis or AAH) seem to maintain that it is possible to explain most of the uniquely human traits as adaptive responses to a specific external factor (e.g., Morgan, 1997 ; Vaneechoutte, Kuliukas, & Verhaegen, 2011 ), but these views have found little resonance in paleoanthropological journals (Bender et al., 2012 ). Indeed, AAH has been fiercely opposed and criticized for being an umbrella hypothesis that attempts to explain everything, for being unparsimonious, for lacking evidence and even for being pseudoscience (Hawks, 2005 ; Langdon, 1997 ; Moore, 2012 ).
Here, we aim to find out what scientists really think about why some of the most striking human traits have emerged. We do so by analyzing the results of an online survey where scientists were directly asked for their views on the issue.
2. MATERIALS AND METHODS
2.1. survey.
A survey was performed using an online form in early 2013. Invitation to participate in the survey was sent by email to the authors of articles and review papers that had been published in a scientific journal of a relevant field during the three previous years (2010–2012). A 3‐year period was thought to be long enough for most researchers to have published at least one scientific paper, but short enough for most of the email addresses given in those papers not to have become obsolete. The focus was on journals of paleontology, zoology, ecology, evolutionary biology, and human biology. Only journals with an ISI impact factor equal to or larger than 1.0 were considered. The exact criteria used to select the journals, as well as a full list of journal names, can be found in Appendix S1 .
Almost 58,000 unique email addresses were found in the information available online for the papers published in the selected journals during the selected time period. The full address list exceeded the capacity of the online survey system (Webropol), so the addresses were sorted in alphabetical order, and an invitation to participate in the survey was sent to the first 29,000 addresses. The remaining addresses were used for a different survey, whose results will be reported elsewhere. The first page of the online survey informed participants about the purpose of the survey. The survey was performed anonymously, and all who responded did so voluntarily. After a few reminders had been sent, a total of 1,266 persons had submitted their responses to the survey.
Although the initial sample was large and can be considered representative of the scientific community in relevant fields, the proportion of invitees who answered the survey was very small (4.4%). The sample is no doubt biased toward people who have a larger than average interest in human evolution. Therefore, the obtained answers do not reflect the opinions of the entire scientific community. Nevertheless, they can indicate whether any of the hypotheses proposed to explain the evolutionary origin of a specific human trait is universally accepted or rejected. Even if this were not the case, the survey gives indication of which hypotheses are most or least popular, although conclusions in this respect remain tentative.
The survey first asked background information of the respondent, such as gender, age, the highest academic degree obtained, number of scientific publications authored (both overall and on human evolution), degree of knowledge about human evolution, and whether the respondent has taught courses on human evolution. The second part listed fifteen human traits (such as bipedalism) and asked the respondents to rate the credibility of 51 alternative hypotheses that have been proposed to explain their evolutionary origin (such as freeing the hands for tool use or seeing over tall grass). The credibility scoring was done using a five‐point scale: very unlikely, moderately unlikely, no opinion, moderately likely, and very likely. The number of alternative hypotheses considered was ten for both bipedalism and brain size, eight for hairlessness, seven for speech, four for subcutaneous fat, and three for descended larynx. In addition, there were nine traits for which only one explanation has been proposed in the literature, and this was related to the aquatic ape hypothesis. The third part asked about the respondents’ views on criticism against AAH. All questions and a summary of the answers are presented in Appendix S2 .
2.2. Data analyses
The respondents were asked for their professional field of expertise by offering 15 alternatives. For statistical analyses, these were simplified to four categories to ensure sufficient sample size in each. The group “(paleo)anthropologist” was formed by lumping the originally separate fields “paleoanthropology” and “anthropology or archaeology.” The group “biologist” was formed by lumping all the original subfields of biology (animal physiology, anatomy, or morphology; ecology; evolution; genetics or molecular biology; other) and the group “human biologist” by lumping all subfields of human biology (cardiovascular or respiratory system, musculoskeletal system, nervous system, nutrition, other aspects of human biology). The fourth group was “other,” which contained the remaining fields (geology, paleontology, other).
Overall relationships among the hypotheses were visualized by principal coordinates analysis (PCoA), where the objects were the hypotheses and the descriptors were individual respondents, with the variable of interest being the credibility score each respondent had given to each hypothesis. A Euclidean distance matrix was calculated, such that the distance between two hypotheses reflects how differently the respondents scored their credibilities. Every respondent who gave one of the hypotheses a higher score than the other increased the final distance between the hypotheses, with the overall distance between the hypotheses equaling zero if every respondent had scored both hypotheses similarly (irrespective of whether the score itself was high or low). PCoA visualizes these pairwise distances, so the closer together two hypotheses get plotted in the ordination diagram, the more similar their explanatory value is in the opinion of an average individual respondent.
The respondents themselves were plotted in the PCoA ordination space on the basis of the scores they had given to the hypotheses. Therefore, the relative positions of the respondents reflect their opinions on the hypotheses: Respondents get plotted toward the same part of the ordination space as the hypotheses they gave highest credibility scores, and far away from the hypotheses they gave lowest scores.
Relationships between the respondents’ opinions and their backgrounds were first assessed visually with the help of the ordination diagram. We then used analysis of variance to test whether there were differences in the average opinions of respondents of different backgrounds. If so, a post hoc Tukey's honest significance test was carried out to assess which aspects of the respondents’ background were associated with differences in opinion. A more detailed breakdown of the respondents’ opinions was obtained by visually comparing the distributions of the credibility scores given to the different hypotheses. This was done both to obtain an idea of which hypotheses are most popular overall, and to see if there were differences among respondents representing different scientific fields and/or having different levels of scientific experience.
R statistical software version 3.3.2 ( https://cran.r-project.org/ ) was used both to run the analyses and to produce the graphs. The vegan package (Oksanen et al., 2015 ) was used for principal coordinates analysis. The survey data and all R code used to manipulate and analyze the data are available at Opasnet web‐workspace http://en.opasnet.org/w/Evolutionary_origin_of_human_traits . The survey data are also available from the Dryad Digital Repository https://doi.org/10.5061/dryad.s9r98 .
Principal coordinates analysis revealed some clear patterns among the hypotheses proposed to explain the evolutionary origin of specific human traits. The most eye‐catching feature of the ordination diagram in Figure 2 a is that the hypotheses got divided into two elongated groups that parallel each other but are clearly separated (the abbreviations of Fig. Fig.2 2 are explained in Table 1 ). The smaller group contains all the hypotheses that evoke adaptation to swimming or diving as an explanatory factor for the emergence of a trait, and the larger group contains all other hypotheses, whether they refer to adaptation to a specific environment or to needs that emerge from a specific behavior. Because all the hypotheses in the smaller group refer to locomotion in water and have been included in the aquatic ape hypothesis (AAH), this group will be referred to as the water‐related or AAH group. For lack of a better unifying term, the larger group will be referred to as the dryland group.

Principal coordinates analysis ( PC oA) of different hypotheses proposed to explain the evolutionary origin of specific human traits. Distances between hypotheses are based on scores given by (a) all respondents, or only respondents whose main field of expertise is (b) anthropology or paleoanthropology, (c) biology, (d) human biology, or (e) other. Each colored point corresponds to one hypothesis, and the color indicates which of the traits listed in the inset the hypothesis aims to explain. Points are scaled to reflect the average credibility score given to the corresponding hypothesis by the respondents of the mentioned expertise group. The hypothesis name abbreviations are explained in Table 1 . Each gray point in (a) corresponds to one respondent, whose position within the ordination space reflects the scores given to the hypotheses. For example, respondents plotted toward the bottom left part of the respondent cloud found the hypotheses plotted toward the bottom left of the hypothesis cloud more credible than the hypotheses at the top, and vice versa. More details on the respondent ordination are shown in Figure 3
The hypotheses on the evolutionary origin of human traits that were included in an online survey to find out how popular they are among scientists. The abbreviations are used in the figures, and the full text is copied verbatim from the survey. If ambiguous, the abbreviated hypothesis is followed by a letter depicting the trait: B = bipedalism, E = encephalization (big brain), F = subcutaneous fat, N = nakedness, L = descended larynx, S = speech, O = other
Within each of the two groups, the hypotheses got sorted by their popularity, with the average credibility score increasing toward the bottom left in Figure 2 a. A tight cluster at the extreme left of the dryland group was formed by five hypotheses with high average credibility scores (4.08–4.26 on a 1–5 scale, with 1 corresponding to “very unlikely” and 5 to “very likely”). This cluster included the most popular hypothesis for the subcutaneous fat layer (energy reserve especially for the developing brain), the descended larynx (required by articulate speech), bipedalism (use of tools and weapons), speech (social pressure for elaborate communication), and the big brain (complex social organization).
This combination might be the most popular overall scenario for the origin of these traits, but the next most popular 2–3 explanations for bipedalism (freeing hands for foraging, better view over tall grass), large brain (required by either language or collaborative hunting), and speech (required by either collaborative hunting or transmitting cultural tradition; triggered by the descended larynx) also received high average credibility scores (3.53–3.96). Their proximity in ordination space indicated that they were found credible by the same respondents, which makes it difficult to identify a single most popular overall scenario. The hypotheses explaining hairlessness were not found convincing by the respondents, as even the two most popular ones (avoidance of overheating when hunting, avoidance of ectoparasites) had average credibility scores of only 3.48 and 3.17, respectively.
Eleven of the twelve most popular hypotheses were based on inherent drivers of evolution, that is, proposing that morphological traits emerged in response to selection pressure either from a novel behavior or from a pre‐existing morphological trait. Hypotheses based on selection pressure from a new kind of external environment were less popular even within the dryland group, and the credibility scores of all the hypotheses in the water‐related group were low to intermediate (2.26–2.99). The hypotheses proposing that encephalization was triggered by improved nutrition also received intermediate popularity scores, whether achieved by cooking or by increased consumption of fish or meat (all three with credibility scores in the range 2.61–2.77). The four least popular hypotheses of all (credibility scores 1.95–2.20) were based on inherent drivers operating on dry land.
The ordination results suggest that the respondents viewed the water‐related hypotheses as an ensemble whose overall credibility they assessed independently of how they scored the credibilities of the other hypotheses. This impression is strengthened when viewing the ordination of the respondents (the gray cloud in Figure 2 a) in more detail (Figure 3 ). The main gradient among the respondents follows the average credibility score they gave for the water‐related hypotheses (Figure 2 a), and this is almost perpendicular to the (less clear) gradient of average credibility scores given for the twelve most popular hypotheses (Figure 3 b).

The positions of the survey respondents in the space of the principal coordinates analysis shown in Figure 2 a. The ordination is the same in each panel, but colors illustrate different kinds of information related to each respondent. The colored crosses indicate the mean position of the respondents belonging to the respective subgroup. (a) Average credibility score given to the hypotheses in the water‐related group (the smaller cloud of points in Figure 2 a). (b) Average score given to the 12 most popular hypotheses in Figure 2 a. (c) Number of scientific publications authored or co‐authored (crosses of all three categories overlap). (d) Field of expertise. (e) Familiarity with hypotheses on human evolution. (f) Experience in teaching human evolution
The respondents’ position in the ordination did not seem to be related with how much scientific experience they had in general, as measured with the total number of scientific publications they had authored (Figure 3 c), but it was related with how much they knew about human evolution. Those having more background information on this specific topic (by self‐assessment, by main field of expertise being paleoanthropology or anthropology, or by having taught university courses on the topic) appeared to be more often plotted in the upper part of the ordination than respondents representing other backgrounds (Figure 3 d–f).
The visual impressions were confirmed by statistical analyses. These were carried out separately for five different subgroupings of the hypotheses. Three of these were chosen because they formed clear groups in the ordination of Figure 2 a (the dryland hypotheses, the water‐related hypotheses, the 12 most popular dryland hypotheses). The dryland hypotheses were also split into those based on environmental adaptation and those evoking behavioral drivers.
The largest effect by far on the responses was that of the field or expertise, with (paleo)anthropologists being more critical overall than representatives of any other expertise group (Table 2 ). The difference was especially large for the water‐related hypotheses: The average credibility score given by (paleo)anthropologists to this group of hypotheses (2.10 on the 1–5 scale) was much lower than the average score given by human biologists (3.02), with biologists (2.70), and others (2.67) being intermediate. For the dryland hypotheses, the difference between (paleo)anthropologists (2.97) and human biologists (3.22) was only 0.25 (vs. 0.92 in the case of the water‐related hypotheses), and the differences in the scores given by biologists, human biologists, and others were not statistically significant.
Results of Tukey's HSD test between different subgroups of respondents (line starting with Test result ~) and their average credibility scores (standard deviation in parentheses) for different groups of hypotheses: the most popular 12 hypotheses; the dryland hypotheses (the larger hypothesis group in Figure 2a); the water‐related hypotheses (the smaller hypothesis group in Figure 2a); dryland hypotheses based on behavioural demands; dryland hypotheses based on adaptation to the external environment
The results obtained with respondent subgroups based on total number of authored peer reviewed publications and total number of authored popular science publications are not shown, because they were not associated with significantly different ( p < .05) means in any comparisons.
*** p < .001; ** p < .01; * p < .05.
Overall scientific experience (as measured with the number of scientific publications authored) had no effect on the scores given to either the dryland or the water‐related hypotheses (Table 2 ). However, the more knowledge the respondents had on human evolution specifically (self‐assessed familiarity with the hypotheses, number of scientific publications on human evolution or experience in teaching human evolution), the lower the scores they gave to the water‐related hypotheses. Among biologists, those who knew more about human evolution were more critical than the less knowledgeable ones, and (paleo)anthropologists were more critical than human biologists with the same self‐assessed knowledge level.
When the dryland hypotheses were split into two groups depending on whether they were based on behavioral arguments or environmental adaptation, both groups obtained rather similar results. The main difference was that the behavioral hypotheses received somewhat higher average credibility scores, which reflects the fact that 10 of the 12 most popular hypotheses were based on behavior (on the other hand, so were the four least popular hypotheses).
To visualize the differences in opinion among the (paleo)anthropologists and representatives of other fields, we repeated the ordination of the hypotheses for each of the four respondent groups separately. In accordance with the fact that most respondents were biologists, the ordination based on the biologists’ data only (Figure 2 c) was very similar to the ordination based on all respondents (Figure 2 a). The ordination based on (paleo)anthropologists’ views (Figure 2 b) differed especially in relation to the hypotheses for bipedalism: Hypotheses that explained bipedalism by foraging, tool use, or carrying were very far removed from the main cloud and toward the opposite side than the water‐related hypotheses. In addition, the average credibility scores given to the water‐related hypotheses were among the lowest of any hypotheses. This contrasted with the situation in the ordination based on human biologists’ data (Figure 2 d), in which the water‐based hypotheses had intermediate credibility scores.
The hypotheses differed clearly from each other in the frequencies of different credibility scores, but there were some similarities in the overall pattern among those six traits for which three or more hypotheses were evaluated (Figure 4 ). None of the hypotheses received the “very likely” score from more than 46% of the respondents, but most traits had at least one hypothesis that was considered “very likely” by more than 23% and likely (either “very likely” or “moderately likely”) by 72%–90%. Many of the intermediately popular hypotheses divided the respondents rather evenly between those who found them likely and those who found them unlikely (the latter referring to the scores “very unlikely” and “moderately unlikely” combined).

Credibility scores given by survey respondents to hypotheses that aim to explain the evolutionary origin of specific human traits. The hypotheses are sorted in order of decreasing popularity as estimated by the percentage of respondents who scored them likely (i.e., either “very likely” or “moderately likely”). Descriptions of the hypotheses as they were given in the survey are shown in Table 1
A causal relationship between articulate speech and descended larynx was accepted by most respondents, but there was no consensus on the direction of the causality. That the larynx descended because this was required by articulate speech was found likely by 84% and very likely by 43%. At the same time, that the evolution of speech was triggered by the descended larynx was found likely by 61% and very likely by 18%. In fact, 36% of the respondents scored both directions as equally likely.
Traits in the category “other” had only one explanatory hypothesis each in the survey, and this was water‐related. All of these hypotheses received many more “very unlikely” than “very likely” scores. However, four hypotheses (that baby swimming, profuse sweating, diving ability, and magnitude of diving reflex evolved as adaptations to a semi‐aquatic way of life) received so many “moderately likely” scores that the percentage of respondents who found them likely was slightly larger than the percentage who found them unlikely (Figure 4 ).
Details on how the hypotheses were scored by respondents representing different fields of expertise are shown in Figure 5 . In accordance with the statistical test results, most hypotheses received rather similar scores from respondents of all fields of expertise. However, (paleo)anthropologists were clearly more critical than representatives of the other fields in relation to several hypotheses, including: that nakedness evolved to avoid ectoparasites, that the big brain evolved because warfare caused pressure for higher intelligence, and that any traits evolved as adaptations to swimming or diving.

Frequencies of credibility scores given to hypotheses aiming to explain different traits (columns) by respondents of different fields of expertise (rows). In each panel, the answers are, from left to right, “very likely,” moderately likely,” “no opinion,” “moderately unlikely,” and “very unlikely.” Hypotheses that have been included in the aquatic ape hypothesis are shown in shades of blue and green. Those dryland hypotheses for which the opinions of anthropologists and other expertise groups clearly diverged are shown in magenta. The other hypotheses are in shades of brown, with darker colors given to hypotheses that received higher average credibility scores in the survey
There was a lot of variation among the traits in how many of the proposed explanations the respondents found convincing (Figure 6 ). For any one trait, 33%–64% of the respondents did not find any of the proposed hypotheses “very likely,” while 19%–38% found exactly one and 8%–45% more than one. Ten respondents (0.8%) explained that they did not score any of the hypotheses as likely, because they do not believe that humans have evolved at all (most of them explicitly referred to special creation by God).

The number of hypotheses (colors) proposed to explain each human trait (rows) that each respondent found very likely (left panel) or likely (either very likely or moderately likely; right panel). The total number of hypotheses included in the survey is shown after the name of each trait
The survey asked respondents’ opinions on twenty critical arguments that have been presented against the aquatic ape hypothesis. For most arguments, the modal response was “no opinion,” especially among those 43% of the respondents who had never heard of AAH before. Nevertheless, some arguments were clearly more frequently agreed with than others (Figure 7 and Table 3 ). The most widely accepted critique was that not all aquatic mammals have naked skin, so hairlessness cannot be considered an aquatic adaptation. In the other extreme, less than 3% of the respondents fully agreed and less than 12% mostly agreed with the critique that AAH is unscientific or not worthy of attention for the reasons given; in most cases, the number of respondents who strongly disagreed with these critiques was larger than the number who mostly or fully agreed.

The degree to which respondents representing different expertise fields agree with critique presented against the aquatic ape hypothesis. The full description of each point of critique can be found in Table 3
Points of critique presented against the aquatic ape hypothesis (AAH). The abbreviations are used in Figure 7 , and the full text is copied verbatim from the survey
4. DISCUSSION
The main results of our survey can be summarized as follows: (1) There was no general agreement among the respondents on why any of the uniquely human traits have evolved: None of the proposed hypotheses was universally either accepted or rejected. (2) For any individual trait, the percentage of respondents who found none of the hypotheses “very likely” was between >30% (bipedalism) and >65% (nakedness). (3) In general, opinions on the credibility of the hypotheses were independent of a person's background (gender, age, field of expertise, degree of scientific experience), but (paleo)anthropologists were clearly more critical than representatives of other fields. (4) The hypotheses that mention adaptation to swimming or diving as an explanatory factor were found much less credible by (paleo)anthropologists and slightly more credible by human biologists than by biologists and representatives of other fields. (5) Most respondents were critical about the aquatic ape hypothesis (AAH), but only a small minority considered it to be unscientific.
Of course, all conclusions based on the survey data must be considered tentative only, because the response rate was very low, and it is possible that the results are biased. Members of some subgroup might have been more likely to respond than members of some other subgroup, and the average credibility scores given to the different hypotheses by the respondents may not be representative of the opinions of all scientists in the background population. However, it is unlikely that a lack of general agreement on the drivers of trait evolution or such a clear difference in opinion between (paleo)anthropologists and others could have emerged just as a result of biased sampling.
Our results did not reveal a set of explanations that would collectively provide a coherent and popular scenario for the origin of all (or even many) human traits. Indeed, some of the hypotheses that had almost equal and rather high average credibility scores explained the same trait, whereas for other traits, no hypothesis emerged as particularly popular. Against this background, it is interesting that almost half of the respondents fully or mostly agreed with the statement that the aquatic ape hypothesis “is not needed, because all human traits can be explained by terrestrial scenarios”.
The lack of agreement on why humans evolved the traits we have today is very obvious in our results: No hypothesis was universally accepted, and for most traits, there were several almost equally popular alternative hypotheses rather than one that would generally be considered superior to the others. None of the hypotheses received the score “very likely” from more than half of the respondents or obtained an average credibility score higher than 4.26 (of 5). For hairlessness, the most popular hypothesis was thought to be “very likely” by only 16% of the respondents, and its average credibility score (3.48) was closer to 3 (which is the limit between being considered more likely than unlikely) than to 4 (moderately likely). In addition, for only two of the traits (subcutaneous fat layer and descended larynx), the most popular hypothesis was found at least moderately likely by almost all respondents at the same time as the next most popular hypothesis was found clearly less likely. This may partly reflect the fact that fewer alternative hypotheses have been proposed for these traits than for many of the others included in the survey.
Importantly, lack of agreement did not reflect just ignorance on the topic among nonspecialists, because the responses were, in general, very similar between anthropologists and respondents representing other fields of science. In fact, anthropologists were even more skeptical about all hypotheses than representatives of the other fields were. In other words, outsiders were slightly more convinced that the proposed hypotheses are plausible than those who work in the field. Maybe anthropologists (especially paleoanthropologists) are more systematically trained to be wary of just‐so‐stories (explanations of past events and processes backed up by little or no evidence) than students in nearby fields are. It is also possible that outsiders are somewhat less likely to question hypotheses proposed within an unfamiliar field. This could be because they do not feel qualified to do so, or because they have not heard of the debates that draw attention to the weaknesses of the hypotheses.
Our results conform with the widespread belief that professionals in the field of human evolution are more critical toward the aquatic ape hypothesis (AAH) than outsiders are (Langdon, 1997 ; Bender et al., 2012 ; see also nonscientific sources such as Hawks, 2005 ; Moore, 2012 and Wikipedia: Aquatic Ape Hypothesis: Talk). However, this did not seem to be due to overall scientific ignorance, because how respondents assessed the credibility of the hypotheses proposing adaptation to swimming or diving was independent of both their overall scientific experience level and how they assessed the credibility of the other hypotheses. Interestingly, those whose main field of expertise is human biology had the most positive attitudes toward the water‐related hypotheses, giving them an average credibility score that was as much as 0.9 units higher (on a 1–5 scale) than the average score given by (paleo)anthropologists.
The difference in average opinion between (paleo)anthropologists and other scientists can be interpreted in two opposite ways. On the one hand, those who know the field of human evolution best may be best positioned to make a justified evaluation of the validity of the alternative hypotheses. On the other hand, prior knowledge may induce one to reject unconventional hypotheses offhand merely because they challenge the established paradigms of a field (Bender et al., 2012 ; Klayman, 1995 ). Obviously, the two interpretations lead to opposite conclusions on whether or not the critical attitude of the (paleo)anthropologists can be taken as evidence that AAH is flawed. In our survey, a vast majority of the respondents who had an opinion on the issue disagreed with the statement that AAH can be ignored because its main proponents are not professionals in the field of human evolution. This was the case both overall and within each field of expertise separately, although the proportion of respondents who agreed with the statement was higher among (paleo)anthropologists than among representatives of the other fields.
In this context, it is also interesting that the respondents’ assessment of the credibility of the water‐related hypotheses did not depend on the number of scientific papers they had authored. This indicates that established scientists are no more likely to reject or accept these hypotheses than junior scientists are—unless their scientific experience relates directly to the field of human evolution. A vast majority of the respondents disagreed with the critique that AAH is unscientific. Of course, this does not mean that they would consider the explanations proposed by AAH to be correct, and indeed, all the hypotheses related to AAH received relatively low credibility scores (although not as low as the least popular dryland hypotheses).
If, for the sake of argument, we accept the most popular explanation for each trait to be the correct one, a scenario of evolution by internal drive emerges: The large brain evolved because complex social organization required higher intelligence, the subcutaneous fat layer evolved to serve as an energy reserve for the developing brain, articulate speech evolved because there was social pressure for elaborate communication, the larynx descended because this was required by articulate speech, bipedalism evolved to make the use of tools and weapons easier, and nakedness evolved to avoid overheating when hunting. For most traits, the next most popular explanation was not far behind in popularity. Most of these were also based on inherent drivers, but sometimes in the opposite temporal sequence (e.g., articulate speech was triggered by the descended larynx; large brain evolved because it was required by articulate speech). We found this result disturbing, because the overwhelming popularity of hypotheses based on inherent drivers gives the impression that human evolution is generally thought to have been goal‐directed. This would be in conflict with the current understanding (explained in every evolutionary biology textbook) that evolution has no foresight.
Overall, the survey revealed no general agreement among the respondents: None of the proposed hypotheses on why specific uniquely human traits have evolved was universally either accepted or rejected. Nevertheless, identifying and quantifying what is not generally known and agreed upon can be useful in itself, as it may help to focus future research on answering the most important open questions. Clearly, there is still a long way to go before the question “why are humans so different from other primates” has been answered in a comprehensive and generally satisfactory way.
DATA ACCESSIBILITY
Conflict of interest.
None declared.
AUTHOR CONTRIBUTIONS
HT designed and conducted the survey and led the writing. All authors discussed the results and planned the data analyses together. The R code used to analyze the data and draw the figures was written by MT with contributions from JT.
Supporting information
Acknowledgments.
We thank Carlos Peña for writing the code to extract respondents’ email addresses from the Internet; Mirkka Jones, Kalle Ruokolainen, and Timo Vuorisalo for comments that helped to improve the survey questions; and Jouko Tuomisto for comments on the manuscript.
Tuomisto H, Tuomisto M, Tuomisto JT. How scientists perceive the evolutionary origin of human traits: Results of a survey study . Ecol Evol . 2018; 8 :3518–3533. https://doi.org/10.1002/ece3.3887 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Barber, N. (1995). The evolutionary psychology of physical attractiveness: Sexual selection and human morphology . Ethology and Sociobiology , 16 , 395–424. https://doi.org/10.1016/0162-3095(95)00068-2 [ Google Scholar ]
- Bartholomew, G. A. , & Birdsell, J. B. (1953). Ecology and the Protohominids* . The American Anthropologist , 55 , 481–498. [ Google Scholar ]
- Bender, R. , Tobias, P. W. , & Bender, N. (2012). The savannah hypotheses: Origin, reception and impact on paleoanthropology . History & Philosophy of the Life Sciences , 34 , 147–184. [ PubMed ] [ Google Scholar ]
- Carrier, D. R. , Kapoor, A. K. , Kimura, T. , Nickels, M. K. , Scott, E. C. , So, J. K. , & Trinkaus, E. (1984). The energetic paradox of human running and hominid evolution [and comments and reply] . Current Anthropology , 25 , 483–495. https://doi.org/10.1086/203165 [ Google Scholar ]
- Crompton, R. H. , Sellers, W. I. , & Thorpe, S. K. S. (2010). Arboreality, terrestriality and bipedalism . Philosophical Transactions of the Royal Society B: Biological Sciences , 365 , 3301–3314. https://doi.org/10.1098/rstb.2010.0035 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cunnane, S. C. , & Crawford, M. A. (2014). Energetic and nutritional constraints on infant brain development: Implications for brain expansion during human evolution . Journal of Human Evolution , 77 , 88–98. https://doi.org/10.1016/j.jhevol.2014.05.001 [ PubMed ] [ Google Scholar ]
- Dart, R. (1925). Australopithecus africanus : The man‐ape of South Africa . Nature , 115 , 195–199. https://doi.org/10.1038/115195a0 [ Google Scholar ]
- Domínguez‐Rodrigo, M. (2014). Is the “Savanna Hypothesis” a dead concept for explaining the emergence of the earliest hominins? Current Anthropology , 55 , 59–81. https://doi.org/10.1086/674530 [ Google Scholar ]
- Gee, H. (2013). The accidental species: Misunderstandings of human evolution . Chicago, IL: University of Chicago Press; https://doi.org/10.7208/chicago/9780226044989.001.0001 [ Google Scholar ]
- Giles, J. (2011). Naked love: The evolution of human hairlessness . Biology Theory , 5 , 326–336. [ Google Scholar ]
- Hardy, A. (1960). Was man more aquatic in the past? The New Scientist , 7 , 642–645. [ Google Scholar ]
- Hawks, J. (2005). Why anthropologists don't accept the Aquatic Ape Theory . Retrieved from http://johnhawks.net/weblog/topics/pseudoscience/aquatic_ape_theory.html [accessed 5 September 2017]
- Hewes, G. W. (1961). Food transport and the origin of hominid bipedalism . The American Anthropologist , 63 , 687–710. https://doi.org/10.1525/aa.1961.63.4.02a00020 [ Google Scholar ]
- Hunt, K. D. (1994). The evolution of human bipedality: Ecology and functional morphology . Journal of Human Evolution , 26 , 183–202. https://doi.org/10.1006/jhev.1994.1011 [ Google Scholar ]
- Hunt, K. D. (1996). The postural feeding hypothesis: An ecological model for the evolution of bipedalism . South African Journal of Science , 92 , 77–90. [ Google Scholar ]
- Isler, K. , & Van Schaik, C. P. (2014). How humans evolved large brains: Comparative evidence . Evolutionary Anthropology: Issues, News, and Reviews , 23 , 65–75. https://doi.org/10.1002/evan.21403 [ PubMed ] [ Google Scholar ]
- Jolly, C. J. (1970). The seed‐eaters: A new model of hominid differentiation based on a baboon analogy . Man , 5 , 5–26. https://doi.org/10.2307/2798801 [ Google Scholar ]
- Kingdon, J. (2003). Lowly origin: Where, when, and why our ancestors first stood up . Princeton, NJ: Princeton University Press. [ Google Scholar ]
- Kingston, J. D. (2007). Shifting adaptive landscapes: Progress and challenges in reconstructing early hominid environments . The American Journal of Physical Anthropology , 134 , 20–58. https://doi.org/10.1002/(ISSN)1096-8644 [ PubMed ] [ Google Scholar ]
- Klayman, J. (1995). Varieties of confirmation bias In Busemeyer J., Hastie R., & Medin D. L. (Eds.), Psychology of learning and motivation (pp. 385–418). Cambridge, MA: Academic Press. [ Google Scholar ]
- Kovarovic, K. , & Andrews, P. (2007). Bovid postcranial ecomorphological survey of the Laetoli paleoenvironment . Journal of Human Evolution , 52 , 663–680. https://doi.org/10.1016/j.jhevol.2007.01.001 [ PubMed ] [ Google Scholar ]
- Kuliukas, A. (2002). Wading for food the driving force of the evolution of bipedalism? Nutrition and Health , 16 , 267–289. https://doi.org/10.1177/026010600201600402 [ PubMed ] [ Google Scholar ]
- Langdon, J. H. (1997). Umbrella hypotheses and parsimony in human evolution: A critique of the Aquatic Ape Hypothesis . Journal of Human Evolution , 33 , 479–494. https://doi.org/10.1006/jhev.1997.0146 [ PubMed ] [ Google Scholar ]
- Leakey, R. , & Lewin, R. (1977). Origins: The emergence and evolution of our species and its possible future . Boston, MA: E. P. Dutton. [ Google Scholar ]
- Lovejoy, C. O. (1981). The origin of man . Science , 211 , 341–350. https://doi.org/10.1126/science.211.4480.341 [ PubMed ] [ Google Scholar ]
- Maslin, M. A. , & Christensen, B. (2007). Tectonics, orbital forcing, global climate change, and human evolution in Africa: Introduction to the African paleoclimate special volume . Journal of Human Evolution , 53 , 443–464. https://doi.org/10.1016/j.jhevol.2007.06.005 [ PubMed ] [ Google Scholar ]
- Moore, J. (2012). Aquatic ape theory: Sink or swim? Retrieved from www.aquaticape.org [accessed 5 September 2017]
- Morgan, E. (1982). The aquatic ape: A theory of human evolution . London, UK: Souvenir Press. [ Google Scholar ]
- Morgan, E. (1990). The scars of evolution: What our bodies tell us about human origins . London, UK: Souvenir Press. [ Google Scholar ]
- Morgan, E. (1997). The aquatic ape hypothesis . London, UK: Souvenir Press. [ Google Scholar ]
- Niemitz, C. (2010). The evolution of the upright posture and gait—a review and a new synthesis . Naturwissenschaften , 97 , 241–263. https://doi.org/10.1007/s00114-009-0637-3 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , Minchin, P. R. , O'Hara, R. B. , … Wagner, H. (2015). Vegan: Community ecology package . R package version 2.3‐2. http://CRAN.R-project.org/package=vegan
- Pontzer, H. , Raichlen, D. A. , & Sockol, M. D. (2009). The metabolic cost of walking in humans, chimpanzees, and early hominins . Journal of Human Evolution , 56 , 43–54. https://doi.org/10.1016/j.jhevol.2008.09.001 [ PubMed ] [ Google Scholar ]
- Potts, R. (1998a). Environmental hypotheses of hominin evolution . The American Journal of Physical Anthropology , 107 , 93–136. https://doi.org/10.1002/(ISSN)1096-8644 [ PubMed ] [ Google Scholar ]
- Potts, R. (1998b). Variability selection in hominid evolution . Evolutionary Anthropology: Issues, News, and Reviews , 7 , 81–96. https://doi.org/10.1002/(ISSN)1520-6505 [ Google Scholar ]
- Ravey, M. (1978). Bipedalism: An early warning system for Miocene Hominoids . Science , 199 , 372 https://doi.org/10.1126/science.199.4327.372 [ PubMed ] [ Google Scholar ]
- Rodman, P. S. , & McHenry, H. M. (1980). Bioenergetics and the origin of hominid bipedalism . The American Journal of Physical Anthropology , 52 , 103–106. https://doi.org/10.1002/(ISSN)1096-8644 [ PubMed ] [ Google Scholar ]
- Stout, D. , & Chaminade, T. (2012). Stone tools, language and the brain in human evolution . Philosophical Transactions of the Royal Society B: Biological Sciences , 367 , 75–87. https://doi.org/10.1098/rstb.2011.0099 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sutou, S. (2012). Hairless mutation: A driving force of humanization from a human–ape common ancestor by enforcing upright walking while holding a baby with both hands . Genes to Cells , 17 , 264–272. https://doi.org/10.1111/j.1365-2443.2012.01592.x [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Tanner, N. M. (1981). On becoming human . Cambridge, UK: CUP Archive. [ Google Scholar ]
- Thorpe, S. K. S. , Holder, R. L. , & Crompton, R. H. (2007). Origin of human bipedalism as an adaptation for locomotion on flexible branches . Science , 316 , 1328–1331. https://doi.org/10.1126/science.1140799 [ PubMed ] [ Google Scholar ]
- Vaneechoutte, M. , Kuliukas, A. , & Verhaegen, M. (2011). Was man more aquatic in the past? Fifty years after Alister hardy – Waterside hypotheses of human evolution . Sharjah, UAE: Bentham Science Publishers. [ Google Scholar ]
- Verhaegen, M. , Puech, P.‐F. , & Munro, S. (2002). Aquarboreal ancestors? Trends in Ecology & Evolution , 17 , 212–217. https://doi.org/10.1016/S0169-5347(02)02490-4 [ Google Scholar ]
- Washburn, S. L. (1960). Tools and human evolution . Scientific American , 203 , 62–75. https://doi.org/10.1038/scientificamerican0960-62 [ PubMed ] [ Google Scholar ]
- Watson, J. C. , Payne, R. C. , Chamberlain, A. T. , Jones, R. K. , & Sellers, W. I. (2008). The energetic costs of load‐carrying and the evolution of bipedalism . Journal of Human Evolution , 54 , 675–683. https://doi.org/10.1016/j.jhevol.2007.10.004 [ PubMed ] [ Google Scholar ]
- Wells, J. C. K. (2006). The evolution of human fatness and susceptibility to obesity: An ethological approach . Biological Reviews , 81 , 183–205. https://doi.org/10.1017/S1464793105006974 [ PubMed ] [ Google Scholar ]
- Wheeler, P. E. (1984). The evolution of bipedality and loss of functional body hair in hominids . Journal of Human Evolution , 13 , 91–98. https://doi.org/10.1016/S0047-2484(84)80079-2 [ Google Scholar ]
- Wheeler, P. E. (1991). The influence of bipedalism on the energy and water budgets of early hominids . Journal of Human Evolution , 21 , 117–136. https://doi.org/10.1016/0047-2484(91)90003-E [ Google Scholar ]

Population genetics: past, present, and future
- Published: 18 July 2020
- Volume 140 , pages 231–240, ( 2021 )
Cite this article

- Atsuko Okazaki 1 , 2 ,
- Satoru Yamazaki 3 ,
- Ituro Inoue 4 &
- Jurg Ott ORCID: orcid.org/0000-0002-6188-1388 2
12k Accesses
6 Citations
6 Altmetric
Explore all metrics
We present selected topics of population genetics and molecular phylogeny. As several excellent review articles have been published and generally focus on European and American scientists, here, we emphasize contributions by Japanese researchers. Our review may also be seen as a belated 50-year celebration of Motoo Kimura’s early seminal paper on the molecular clock, published in 1968.
Similar content being viewed by others

Computational Tools for Population Genomics

Molecular Evolution: A Brief Introduction

Population genetics from 1966 to 2016
Avoid common mistakes on your manuscript.
Introduction
In recent years, large amounts of DNA sequencing data have been generated in various projects such as 1000 Genomes (Genomes Project et al. 2010 , 2012 , 2015 ), the ALSPAC database (Fraser et al. 2013 ; Hameed et al. 2017 ), and Icelandic (Gudbjartsson et al. 2015 ), and Japanese populations (Nagasaki et al. 2015 ). Major achievements of these efforts have been as follows: (1) Larger genetic variation is observed within populations than between populations, and (2) each individual harbors large numbers of variants with low allele frequencies. These findings have long ago been predicted by population genetics and evolutionary studies. Therefore, it is instructive to look back at historic achievements in population genetics.
Excellent reviews of population genetics have been written (Chakraborty 2006 ; Charlesworth and Charlesworth 2017 ; Crow 1987 ; Crow and Kimura 1970 ) documenting the development of population genetics from early achievements by Mendel ( 1866 ), Hardy ( 1908 ), and Weinberg ( 1908 ) up to highly sophisticated theoretical developments, mostly by American, British, and Japanese scientists. Here, we review selected aspects of population genetics, genome evolution, and molecular phylogeny with an emphasis on contributions by Japanese researchers.
Historical aspects of population genetics and road to the neutral theory
Darwin’s theory of evolution through selection very well explains changes in time of heritable phenotypes. In the early 1900s, focusing on the evolution of genetic variants in the population, R. A. Fisher, S. Wright, and J. B. S. Haldane made fundamental theoretical contributions to population genetics (Provine 1971 ), Fisher in his 1922 paper (Fisher 1922 ), which was the first to introduce diffusion equations into population genetics, and Haldane in developing in 1927 (Haldane 1927 ) the approximation of change of numbers of copies of very rare mutants by branching processes. Wright ( 1938 ) developed the theory on the effects of genetic drift, that is, random changes in small populations. While his theory was supported only by a minority of scientists in an era when the molecular basis of genes had yet to be proven and the effects of genetic drift were underestimated, Wright’s theory made a great contribution to connecting Mendelian Genetics with the Darwinian theory of evolution.
More recently, it has become apparent that many molecular changes have no effects on phenotypes. Based on Wright’s drift hypothesis and Haldane’s approximation model of an advantageous mutation (Haldane 1927 ), Motoo Kimura ( 1964 ) then developed his neutral theory based on backward diffusion models, which showed the probability of fixation to zero of a variant in the population to be equal to 2 s ( N e / N ), where s is the selection coefficient, N the size of the breeding population, and N e the effective population size.
Mutations and selection are driving forces for evolution. Basically, mutations occur at random DNA bases. Harmful mutations tend to be eliminated within a short period of time and do not contribute to long-term evolution. This process is called negative or purifying selection as opposed to positive selection. Before Kimura ( 1964 ) proposed his neutral theory, there was little notion of neutral variation, although, at about the same time, Lewontin and Hubby ( 1966 ) considered the possibility of neutral mutation as a possible reason for a large amount of variation which they found in electrophoretic mobility. Still, natural selection was the mainstream hypothesis with the idea that advantageous variations in populations are the driving forces for evolution, and deleterious variations are removed in a rapid manner.
At the time, population genetics usually considered two alleles at each gene locus based on the assumption of genes being base pairs. On the other hand, Kimura and Crow ( 1964 ) assumed an infinite allele model (“neutral isoalleles”) and proposed that genetic variation in populations arises as to the balance between mutations and genetic drift. Comparing hemoglobin molecules between different organisms, Kimura ( 1968 ) postulated that amino-acid substitution rates are so high that they can only be explained by neutral mutations. In other words, mutation and random changes in a finite population can maintain considerable variation through random fixation of selectively neutral or nearly neutral mutants. In the light of current knowledge, however, Kimura’s reasoning appears somewhat flawed. For example, he argued that the “cost of natural selection” would be too high otherwise—more consideration has shown that no cost is imposed by beneficial mutations in the absence of environmental deterioration. He also used the total amount of DNA without distinguishing protein-coding regions and non-coding regions. Nonetheless, Kimura’s contributions to population genetics have been tremendous.
Together with the Darwinian selection hypothesis, the neutral theory is one of the two pillars of genome evolution. Thus, ‘survival of the luckiest, and not necessarily of the fittest’ may be a good explanation for the evolution of a great majority of genetic changes (Chakraborty 2006 ). Interestingly, Kimura ( 1969 ) also proposed the “infinite sites model”. In this model, if the mutation rate is low and the effective population size is small ( θ = 4 N e µ « 1), a mutant variant will always appear at a different site in the genome. If so, identity by state at the variant can be regarded as identity by descent, and in this respect, the infinite sites model represents one of the bases for genome-wide association studies using SNPs as genetic markers in unrelated individuals (Sella and Barton 2019 ).
The nearly neutral theory
The evolutionary rate, λ = fμ , in the neutral theory ( f is the proportion of neutral mutations among all mutations in a gene, μ is the mutation rate) disregards mutations favorable to survival and simply classifies other mutations into neutral ( f ) and deleterious (1 − f ) mutations. However, the extent of harmfulness measured by the selection coefficient, s , is a continuous quantity. Based on these ideas, Tomoko Ohta (Ohta 1973 , 1992 , 2002 ), who had built the foundation of the neutral theory with Motoo Kimura, proposed the “nearly neutral” theory, where slightly disadvantageous mutations (attenuated mutations) could persist in the population by chance if the population is small. Thus, according to her publications (Ohta 1973 , 1992 , 2002 ), a substantial fraction of changes is caused by random fixation of nearly neutral changes, a class that includes intermediates between neutral and advantageous, as well as between neutral and deleterious classes, although other population geneticists may disagree with this view (Kondrashov 1995 ; Nei 2005 ).
A difference from the neutral theory is that the nearly neutral theory allows for interactions between (1) genes having occurred through weak natural selection (or weak deleterious selection) and (2) genes without weak natural selections, and for the two types of genes to jointly contribute to evolution by opposing the action of genetic drift (Hurst 2009 ). In the nearly neutral theory, the effect of genetic drift is weakened, and slightly disadvantageous mutations are excluded from a population if the population is extremely large; if a population is small, then slightly disadvantageous mutations are kept (some are even fixed) by the effects of genetic drift. It seems that the structure of very large datasets such as 1000 Genomes or the Exome Sequencing Project 6500 can be explained by the nearly neutral theory, because there is increasing evidence that selection pressure in small populations such as mammals including humans is weaker compared to that in ancestral species, and slightly disadvantageous mutations have been accumulating in populations (Kosiol et al. 2008 ; Nelson et al. 2012 ; Nielsen et al. 2009 ; Tennessen et al. 2012 ).
Evolutionary rate of pseudogenes
In the second half of 1970, accumulated sequencing data confirmed the prediction by King and Jukes ( 1969 ) that mutation rates of synonymous variants are higher than those of non-synonymous variants, which supports the neutral theory. Kimura ( 1977 ) asserted that according to the neutral mutation-random drift hypothesis, most mutant substitutions detected among organisms should be the results of random fixation of selectively neutral or nearly neutral mutations. This conjecture was verified by the analysis of mutation rates of pseudogenes, that is, of genes with sequences similar to normal genes having lost their functions as they were duplicated to another location in the genome, and in the process, their transcription sequences were not preserved. Based on the neutral theory, Takashi Miyata calculated the replacement rates of non-synonymous variants and synonymous variants in nucleotide sequences of several pseudogenes, α and β globin, and compared them with those in their functional counterparts (Miyata and Hayashida 1981 ). Results showed that replacement rates were uniformly the same in different pseudogenes and almost equal to the mutation rate, with no other gene evolving at a faster rate. This observation clearly supported the neutral theory.
Junk DNA, a term publicized by Susumu Ohno ( 1972 ) but rarely used today (see below), contains inter-genic regions, most of which are SINEs ( S hort IN terspersed E lements) and LINEs ( L ong IN terspersed E lements). The term ‘junk DNA’ was mentioned by a few other authors in 1972 and even 9 years earlier in a paper little known to human geneticists (Ehret and De Haller 1963 ), but Ohno’s name tends to be most closely associated with this term.
Evolutionary rates of junk DNA are expected to be similar to those of synonymous mutations and pseudogenes. In mammals, most of the genome regions, likely well more than 90%, are predicted to be junk DNA. Therefore, evolutionary rates of whole genomes can be approximated as being those of junk DNA.
In 2012, the Encyclopedia of DNA elements (ENCODE) project (Consortium 2012 ) proved biochemical functions of 80% of the genome, especially outside of protein-coding regions, which was once considered junk DNA. The findings from the ENCODE project enable us to further explore the function of the human genome.
Genes and genomic duplication
In higher organisms, genomic duplication is known to be extremely important for evolution. Early on, Susumu Ohno proposed that evolution is caused by genomic duplication, which was a visionary idea at a time when large sequencing data were not yet available (Ohno 1970 ). It has been shown empirically and by theoretical considerations that the advantage of creating new copies of genomes (or individual genes) can result in higher fitness. An alternative model explaining genomic duplication is DDC ( D uplication D egeneration C omplementation) (Lynch and Conery 2000 ). In the DDC model, regulatory elements each controlling independent functions are duplicated and random null mutations in the regulatory elements through degeneration lead to sub-functionalization, where the regulatory elements complement each other to achieve the full ancestral repertoires. What is important in the process is that it does not require the help of positive selection, that is, functional diversification. In practice, it has been proposed that the selection of slightly disadvantageous mutations works with the expression level of each gene changing. Therefore, genetic duplication is predicted to proceed in a nearly neutral manner based on mutation pressure and genetic drift. In addition, “concerted evolution” in minisatellites used as markers for hyper-polymorphisms, and in other sequences such as rRNA genes can be explained well by Ohno’s theory (Hillis et al. 1991 ; Jeffreys et al. 1985 ).
Molecular phylogeny
Through evolution, currently, living organisms have descended from common ancestors. Systematic biology seeks to unravel relationships among organisms and to establish evolutionary trees. As every biology student knows, the classical approach to such discoveries is through painstaking analysis of morphological details. Depending on which of these phenotypes are considered most important, different relationships among organisms emerge.
Rather than relying on phenotypes that may or may not be heritable, molecular phylogeny relies on DNA sequences and their comparisons among organisms. Researchers with various backgrounds have made significant contributions to methods of creating phylogenetic trees and the evaluation of phylogenetic relationships. In this field, Joseph Felsenstein almost single-handedly established this field as a special branch of population genetics (Felsenstein 2004 ). For example, he introduced the maximum-likelihood method of establishing phylogenetic trees (Felsenstein 1978 ) (see below). One of his other contributions is the “Felsenstein Zone” (Huelsenbeck and Hillis 1993 ), which involves the phenomenon of “long-branch attraction”; that is, long branches will appear similar to each other and appear as sister taxa on a tree even though they do not share a common ancestry. The Zone is the set of trees on which long-branch attraction occurs. Such phenomena have been observed in many datasets and simulation analyses, and have led to the discovery of long-branch attraction, which leads to wrongly assuming phylogeny where none exists (Huelsenbeck and Hillis 1993 ). Furthermore, Felsenstein contributed greatly to molecular phylogeny by developing a program package, PHYLIP, combining various phylogenic tree estimation methods including DNAML. Thanks to his contributions, molecular phylogeny has become increasingly popular for empirical molecular evolutionists.
The development of molecular phylogeny may not seem to be related to disease gene discovery. However, it greatly contributes to such discoveries through interpretation of huge sequencing datasets obtained from the 1000 Genomes project and other projects. Generating a molecular phylogenetic tree for phylogenetic relationships between species led to the discovery of gene families (orthologs and paralogs). The coalescent theory, which examines the gene tree in a species by reversing the time, was also applied to reconstruct the demographic history of species of interest. In particular, regarding the coalescent theory, Tajima ( 1983 ) estimated nucleotide diversity based on the limited DNA polymorphic data, calculated the time of coalescence of genes sampled from a single population, and their theory applies to a few genes at the time of population splitting. Takahata and Nei ( 1985 ) further developed a coalescent theory from DNA sequencing data and theoretically showed that alleles with deep coalescences are relatively rare.
The neighbor-joining method
Many methods for creating (estimating) phylogenic trees have been developed. Historically, these methods can roughly be classified into two groups, distance matrix methods and character state methods. The former uses a distance matrix and estimates evolutionary distance such as the number of amino-acid substitutions or base substitutions based on all possible pairs of OTUs (Operational Taxonomic Units). This method was first applied to create phylogenic trees in the form of the UPGMA (Unweighted Pair Group Method with Arithmetic mean) method, where clusters of neighboring OTUs are created and connected in a stepwise fashion. The method is used not only for amino-acid or base-pair sequences but also in numerical taxonomy, which deals with expression analysis using microarray (Eisen et al. 1998 ) or trait-encoded information (Sokal and Michener 1958 ). However, since this method assumes constant evolutionary speed, it is problematic to apply to amino-acid or base-pair sequence data. To overcome this problem, distance methods were developed that did not assume a molecular clock (Fitch and Margoliash 1967 ). Masatoshi Nei and Naruya Saitou greatly improved upon this method and developed a much faster procedure (Saitou and Nei 1987 ). This method is one of the “star decomposition” methods that determine which, of a given pair of sequences, reduces length of the total tree most and combine neighboring nodes until all OTUs are included. In the neighbor-joining method, “neighbors” keep track of nodes on a tree rather than taxa or clusters of taxa. A modified distance matrix is obtained in which the separation between each pair of nodes is adjusted on the basis of their average divergence from all other nodes. The tree is constructed by joining the least-distant pair of nodes in this modified matrix. When two nodes are joined, their common ancestral node is added to the tree and the terminal nodes with their respective branches are removed from the tree. At each stage in the process, two terminal nodes are replaced by one new node. This iterative operation finds “neighbors” one after another, which creates the final phylogenetic tree. The neighbor-joining method is the most commonly used distance matrix method. Starting in 1971, Nei proposed that Nei’s distance be used for phylogenetic tree estimation, which was later incorporated into the neighbor-joining program package MEGA (Kumar et al. 1994 ; Saitou and Nei 1987 ).
The second group, character state methods, do not use a distance matrix and define characters (phenotypes) and use them for exploring tree topology. One of the examples of character state methods is the maximum-likelihood method discussed in the next section.
The maximum-likelihood method
Maximum likelihood (ML) was developed by Fisher ( 1922 ) as a method to estimate parameters in statistical models. It has several advantages over other methods, but tends to be more complicated to apply than simpler methods. In population genetics, Luigi Luca Cavalli-Sforza first applied the ML method to an approach for creating phylogenic trees based on allele frequencies (Cavalli-Sforza and Edwards 1967 ). The first use of maximum-likelihood inference of trees from molecular sequences was by Jerzy Neyman (Felsenstein 2001 ; Neyman 1971 ). Felsenstein proposed ML for creating phylogenic trees based on allele frequencies as continuous quantities (Felsenstein 1973a ), thus improving on the method previously proposed by Cavalli-Sforza, and introduced ML for estimating trees based on discrete datasets and the maximum parsimony criterion (Felsenstein 1973b ). Masami Hasegawa incorporated this approach into the MOLPHY program package and pioneered in the use of model selection methods such as AIC in comparing phylogenies (he was a member of Akaike’s institute) (Adachi and Hasegawa 1992 , 1996 ).
The ML method is the most efficient approach among all tree construction methods. For example, false-positive evidence of relationships of long branches (“long-branch attraction”) will not occur when trees are estimated by ML and the model of evolution is correct, although it can occur when the model is not correct. However, the ML method tends to be time-consuming and, for some large trees, may be impossible to apply.
Impact of variants on multifactorial disorders and missing heritability
Based on the material mentioned so far, we will now cover some topics on how progress in population genetics, genome evolution, and phylogenic studies can be applied to medical research.
Multifactorial disorders are assumed to occur through interactions between multiple genetic and environmental factors. Therefore, identifying disease susceptibility genes has been considered difficult, and detecting interactions with environmental factors even more so. Especially in the 1990s, such considerations were widespread, quite in contrast to the relative ease with which increased numbers of gene identifications for monogenic disorders have been achieved. However, there was a researcher to struggle with the solution for genetic causes of multifactorial disorders at that time. Ituro Inoue succeeded in narrowing down disease loci using linkage analysis with affected sib-pairs and constructing haplotypes of the angiotensinogen (AGT) gene using limited data (Inoue et al. 1997 ). Inoue assessed linkage disequilibrium (LD) at each site in the AGT gene and further demonstrated by in vitro functional assay that the combination between A (− 6) and T235 alleles affects the expression of the AGT gene. This study was visionary, since LD block structures had yet to be proved at that time.
After that, genome-wide association studies with large SNP data over the whole genome became available thanks to the HAPMAP project, SNP collections by Perlegen Science, LD block measurements, and construction of haplotype maps (HapMap 2005 ; Hinds et al. 2005 ). Although such genome-wide studies contributed to narrowing down locations of disease susceptibility genes, results are still insufficient for identifying many specific disease susceptibility genes, for example Moyamoya disease (Liu et al. 2011 ). A remaining challenge has been that identified susceptibility loci show only small odds ratios, and all susceptibility loci combined only explain up to 30% of most of the disease causes. These numbers are generally smaller than the heritability calculated in the previous twin studies, which is known as “missing heritability” (Manolio et al. 2009 ). Nowadays, however, methods for calculating SNP-based heritability have been developed (Yang et al. 2017 ) that come up with heritability estimates close to those obtained by classical segregation analysis, and part of the problem seems to be resolved.
Out-of-Africa hypothesis
Recent advances in sequencing technology have enabled the identification of whole genome structures at population levels. These successes have made it possible to compare current human genome sequences with ancient genomes such as Homo neanderthalensis or Denisova hominin , which greatly contributed to the understanding of the origin of Homo sapiens (Nielsen et al. 2017 ). Allan Wilson, along with Rebecca Cann and Mark Stoneking, first proposed the “out-of-Africa” hypothesis (Cann et al. 1987 ), which claims that Homo sapiens originated in Africa and then spread all over the world. They based their results on the analysis of mitochondrial DNA of various populations, which represented the first phylogenic tree of Homo sapiens . Work by Masatoshi Nei contributed to the out-of-Africa hypothesis: In the 1970s, Nei calculated heterozygosity for various protein isozymes and created phylogenic trees of Homo sapiens (Nei and Roychoudhury 1972 , 1974 ; Nielsen et al. 2017 ). An interesting finding based on this work is that genetic variation estimated by Nei’s distance or Wright’s F st is larger within populations than between populations (Lewontin 1972 ), which was later confirmed by the 1000 Genomes project. In other words, there are greater differences among individuals in a given population than between populations. However, this notion has also been challenged (Edwards 2003 ).
Relationship between recent explosive population growth and origin of deleterious variants
Numerous human genome sequence projects such as 1000 Genomes revealed that each individual harbors considerable numbers of private mutations. This fact had been proposed by Haldane in his “genetic load” theory, which predicted an association between the numbers of variants possessed over populations and survival rate (Haldane 1937 ). In his theory, he claimed that if we consider genetic load for the whole genome rather than a given locus, the fitness decrease by mutations is equal to the mutation rate, v , irrespective of the extent of selection. He also claimed that pathogenic mutations accumulate in the form of heterozygous variants unless such mutations are excluded as lethal homozygous mutations (Haldane 1937 ) (this theory is also known as the Haldane–Muller principle). The theory of genetic load was further elaborated upon by Kimura ( 1960 ); for neutral mutations, there is no load. Based on this background, for variants whose distributions differ among populations, estimating the age of each variant becomes possible, which is important for understanding the history of human evolution, as well as for developing novel methods for disease gene discovery. The mathematical theory of coalescence allowing haplotype and allele ages to be calculated was developed by John Kingman ( 2000 ), and Kimura and Ohta ( 1973 ) proposed a formula for determining allele age, − 2 x (1 − x )/log( x ). This formula represents the expected age of a neutral mutation of frequency x in a stationary population based on a diffusion process used in classical population genetics. Although there was a discussion regarding the restrictive assumption that the age distribution of a mutant allele with population frequency x should be the same as the distribution of the time to extinction of the allele, conditional on extinction, it made a great contribution to later calculations of allele age (Fu et al. 2013 ). Calculating allele age assuming the infinite many sites of model of mutation developed Kimura and Ohta formula, it showed that about three-quarters of all protein-coding SNV predicted to be deleterious across in the past 5000 years (Fu et al. 2013 ). This attempt provides important practical information that can be prioritized variants in disease gene discovery.
Inbreeding (mating between relatives) has so far not been discussed here as it does not lead to changes in allele frequencies. It does, however, lead to a decrease in heterozygotes and a corresponding increase in homozygotes. As is well known, at a bi-allelic locus with allele frequency p , the proportion of heterozygotes is given by 2 p (1 − p )(1 − F ), where F is the inbreeding coefficient. In many human populations, F tends to be rather small; for example, F = 0.00038 in the UK (Pattison 2016 ). An exception is offspring of first cousins ( F = 1/16). For rare deleterious recessive traits with disease allele frequency p , recessive offspring of first-cousin marriages occur with probability p 2 + p (1 − p ) F (Haldane and Moshinsky 1939 ). Through genetic linkage of such a trait with SNPs surrounding it, rare recessive traits tend to be located in long runs of homozygous SNPs (homozygosity mapping (Lander and Botstein 1987 )). More modern approaches have been developed, for example, based on the Hamming distance between chromosomes in affected and control individuals (Imai et al. 2015 ). This approach revealed a mutation, p.H96R in the BOLA3 gene, possibly having originated in a single Japanese founder individual (Imai et al. 2016 ).
Darwinian (evolutionary) medicine
From the viewpoint of Darwinian medicine (or evolutionary medicine), which is medicine based on evolution (Williams and Nesse 1991 ), we discuss a few aspects of how discovering variants can translate into medical care.
In the 1960s, Richard Lewontin discovered in Drosophila populations that heterozygosity is more often observed than expected (Lewontin and Hubby 1966 ). He interpreted this finding as advantageous fitness of heterozygosity compared to the homozygous state of the wild type or mutant (so-called over-dominance, or balancing selection) and emphasized its importance for survival. After the establishment of the neutral theory, as described below, the importance of balancing selection for some types of variants with high allele frequencies was rediscovered. Theoretical studies on natural selection also greatly progressed and “Tajima’s D”, developed by Fumio Tajima, is computed as the difference between two measures of genetic diversity: the mean number of pairwise differences and the number of segregating sites, each scaled so that they are expected to be the same in a neutrally evolving population of constant size. This is a unique contribution to statistical genetics by Japanese researchers in that this method can assess whether a given variant scattered over the whole genome is neutral or under selection pressure (Tajima 1989 ).
Analyzing genome sequences in several populations using the techniques of next-generation sequencing reveals some signals with positive selection pressure. One such example is infection-related diseases. Regarding the natural selection for resistance of a pathogen, this was revealed by next-generation sequencing to represent the strongest positive selection pressure in human evolution; that is, the well-known balancing signals on glycoproteins and positive selection signals on TLRs (Ferrer-Admetlla et al. 2008 ). Applying the history of evolution for various pathogens to disease susceptibility research will likely identify functional variants as well as intra-cellular mechanisms and treatment for various diseases. We believe that selection pressure for ancient pathogens will affect not only infectious and auto-immune diseases but also other traits. Recently, the association between life-style diseases and natural selection has become an attractive topic. Using 40 traits from the UK Biobank, functional low-frequency variants have been revealed to be under negative selection (Gazal et al. 2018 ). An alternative suggestion has been that positive selection acts on susceptibility loci for life-style diseases. An example is the thrifty gene hypothesis. At the dawn of the era of genomic medicine, the ancient history of human evolution is a powerful tool for understanding human biology leading to improving human health.
In this outline, we deliberately emphasized contributions to population genetics by Japanese researchers—in this field, Japanese scientists have arguably carried out comprehensive fundamental work. Thus, we feel justified in presenting this short review of population genetics from a Japanese point of view.
In terms of future developments in population genetics, we expect DNA sequencing to play an ever-increasing role. In an era where human genome sequence projects are underway around the world, established population genetics principles will be applied to reveal more detailed migration history, population history, and mechanisms of selection pressure, particularly in small ethnic populations (Antonio et al. 2019 ; Lipson et al. 2020 ).
Technological advances have changed the landscape of genetic screening (Ceyhan-Birsoy et al. 2019 ). Together with epidemiological and molecular genetics studies, population genetics approaches have demonstrated the association between disease mechanisms and mutations in populations. Cystic fibrosis is one such successful example (Bell et al. 2020 ). By identifying the relationship between specific mutations and a cystic fibrosis transmembrane conductance regulator (CFTR) defect, we can improve patient care including disease monitoring and treatment decisions. In the future, improvement of patient care in more diseases can be achieved by the combination of population genetics, epidemiological studies, and molecular genetics studies.
With the huge amount of genomic information currently available, it is challenging to link genotypes to phenotypes, predict regulatory functions, and classify mutant types. Therefore, new and innovative approaches are needed for further understanding of medical biology and connections to genetic disease. One approach is to collect previously reported SNV information and create a suitable mathematical model. As an example, a study by Davis et al. ( 2016 ) describes a biophysical metric of cardiomyocyte function, which accurately predicts human cardiac phenotypes.
Another approach is based on neural networks to automatically extract relevant features from input data (Zou et al. 2019 ). Since advances in sequencing technologies provide large amounts of data, it is realistic to utilize machine learning as a tool for analysis in the field of clinical healthcare and population genetics. Although deep learning has great potential, attempts to apply it to genomics have only just begun. For example, SpliceAI, a 32-layer deep neural network (DNN) was developed for predicting de novo mutations with predicted splice-altering consequences in patients with neurodevelopmental disorders, which paves the way for the application of deep learning on complex genetic variant prediction (Jaganathan et al. 2019 ). To identify pathogenic mutations in patients with rare diseases, a DNN model was developed combining common variants derived from human and six non-human primate species. The proposed model achieved an 88% accuracy and found 14 unreported candidate genes associated with intellectual disability (Sundaram et al. 2018 ).
Finally, epidemics and pandemics of viruses and their sequences provide rich sources of information. For example, population genetic analyses of 103 SARS-CoV-2 genomes indicated the presence of two major lineages, although the implications of these evolutionary changes remained unclear (Tang et al. 2020 ).
Adachi J, Hasegawa M (1992) MOLPHY, programs for molecular phylogenetics. I, PROTML, maximum likelihood inference of protein phylogeny. Computer science monographs, no. 27. Institute of Statistical Mathematics, Tokyo, pp 1–14
Adachi J, Hasegawa M (1996) MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Computer science monographs, no. 28. Institute of Statistical Mathematics, Tokyo, pp 1–150
Antonio ML, Gao Z, Moots HM, Lucci M, Candilio F, Sawyer S, Oberreiter V, Calderon D, Devitofranceschi K, Aikens RC, Aneli S, Bartoli F, Bedini A, Cheronet O, Cotter DJ, Fernandes DM, Gasperetti G, Grifoni R, Guidi A, La Pastina F, Loreti E, Manacorda D, Matullo G, Morretta S, Nava A, Fiocchi Nicolai V, Nomi F, Pavolini C, Pentiricci M, Pergola P, Piranomonte M, Schmidt R, Spinola G, Sperduti A, Rubini M, Bondioli L, Coppa A, Pinhasi R, Pritchard JK (2019) Ancient Rome: a genetic crossroads of Europe and the Mediterranean. Science 366:708–714. https://doi.org/10.1126/science.aay6826
Article CAS PubMed PubMed Central Google Scholar
Bell SC, Mall MA, Gutierrez H, Macek M, Madge S, Davies JC, Burgel PR, Tullis E, Castanos C, Castellani C, Byrnes CA, Cathcart F, Chotirmall SH, Cosgriff R, Eichler I, Fajac I, Goss CH, Drevinek P, Farrell PM, Gravelle AM, Havermans T, Mayer-Hamblett N, Kashirskaya N, Kerem E, Mathew JL, McKone EF, Naehrlich L, Nasr SZ, Oates GR, O'Neill C, Pypops U, Raraigh KS, Rowe SM, Southern KW, Sivam S, Stephenson AL, Zampoli M, Ratjen F (2020) The future of cystic fibrosis care: a global perspective. Lancet Respir Med 8:65–124. https://doi.org/10.1016/S2213-2600(19)30337-6
Article CAS PubMed Google Scholar
Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36
CAS PubMed Google Scholar
Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19:233–257
CAS PubMed PubMed Central Google Scholar
Ceyhan-Birsoy O, Murry JB, Machini K, Lebo MS, Yu TW, Fayer S, Genetti CA, Schwartz TS, Agrawal PB, Parad RB, Holm IA, McGuire AL, Green RC, Rehm HL, Beggs AH, BabySeq Project T (2019) Interpretation of Genomic Sequencing Results in Healthy and Ill Newborns: Results from the BabySeq Project. Am J Hum Genet 104:76–93. https://doi.org/10.1016/j.ajhg.2018.11.016
Chakraborty R (2006) Population Genetics: Historical Aspects. eLS. Wiley, Chichester, pp 1–3
Google Scholar
Charlesworth B, Charlesworth D (2017) Population genetics from 1966 to 2016. Heredity (Edinb) 118:2–9. https://doi.org/10.1038/hdy.2016.55
Article CAS Google Scholar
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. https://doi.org/10.1038/nature11247
Crow JF (1987) Population genetics history: a personal view. Annu Rev Genet 21:1–22. https://doi.org/10.1146/annurev.ge.21.120187.000245
Crow JF, Kimura M (1970) An introduction to population genetics theory. Harper & Row, New York
Davis J, Davis LC, Correll RN, Makarewich CA, Schwanekamp JA, Moussavi-Harami F, Wang D, York AJ, Wu H, Houser SR, Seidman CE, Seidman JG, Regnier M, Metzger JM, Wu JC, Molkentin JD (2016) A tension-based model distinguishes hypertrophic versus dilated cardiomyopathy. Cell 165:1147–1159. https://doi.org/10.1016/j.cell.2016.04.002
Edwards AW (2003) Human genetic diversity: Lewontin's fallacy. BioEssays 25:798–801. https://doi.org/10.1002/bies.10315
Ehret CF, De Haller G (1963) Origin, development and maturation of organelles and organelle systems of the cell surface in Paramecium. J Ultrastruct Res 23:1–42
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868. https://doi.org/10.1073/pnas.95.25.14863
Felsenstein J (1973a) Maximum-likelihood estimation of evolutionary trees from continuous characters. Am J Hum Genet 25:471–492
Felsenstein J (1973b) Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst Biol 22:240–249
Felsenstein J (1978) The number of evolutionary trees. Syst Biol 27:27–33. https://doi.org/10.2307/2412810
Article Google Scholar
Felsenstein J (2001) Taking variation of evolutionary rates between sites into account in inferring phylogenies. J Mol Evol 53:447–455. https://doi.org/10.1007/s002390010234
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, Sunderland
Ferrer-Admetlla A, Bosch E, Sikora M, Marques-Bonet T, Ramirez-Soriano A, Muntasell A, Navarro A, Lazarus R, Calafell F, Bertranpetit J, Casals F (2008) Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol 181:1315–1322. https://doi.org/10.4049/jimmunol.181.2.1315
Fisher RA (1922) On the mathematical foundations of theoretical statistics. Phil Trans Roy Soc A202:309–368
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 155:279–284. https://doi.org/10.1126/science.155.3760.279
Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, Ring S, Nelson SM, Lawlor DA (2013) Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol 42:97–110. https://doi.org/10.1093/ije/dys066
Article PubMed Google Scholar
Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project NES, Akey JM (2013) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–220. https://doi.org/10.1038/nature11690
Gazal S, Loh P-R, Finucane HK, Ganna A, Schoech A, Sunyaev S, Price AL (2018) Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat Genet 50:1600–1607
Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526:68–74. https://doi.org/10.1038/nature15393
Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E, Sigurdsson GT, Stacey SN, Frigge ML, Holm H, Saemundsdottir J, Helgadottir HT, Johannsdottir H, Sigfusson G, Thorgeirsson G, Sverrisson JT, Gretarsdottir S, Walters GB, Rafnar T, Thjodleifsson B, Bjornsson ES, Olafsson S, Thorarinsdottir H, Steingrimsdottir T, Gudmundsdottir TS, Theodors A, Jonasson JG, Sigurdsson A, Bjornsdottir G, Jonsson JJ, Thorarensen O, Ludvigsson P, Gudbjartsson H, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Arnar DO, Magnusson OT, Kong A, Masson G, Thorsteinsdottir U, Helgason A, Sulem P, Stefansson K (2015) Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 47:435–444. https://doi.org/10.1038/ng.3247
Haldane J (1927) A mathematical theory of natural and artificial selection, Part V: selection and mutation. Math Proc Cambridge Philos Soc 23:838–844. https://doi.org/10.1017/S0305004100015644
Haldane JBS (1937) The effect of variation on fitness. Am Nat 71:337–349
Haldane JBS, Moshinsky P (1939) Inbreeding in mendelian populations with special reference to human cousin marriage. Ann Eugen 9:321–340
Hameed MA, Lingam R, Zammit S, Salvi G, Sullivan S, Lewis AJ (2017) Trajectories of early childhood developmental skills and early adolescent psychotic experiences: findings from the ALSPAC UK birth cohort. Front Psychol 8:2314. https://doi.org/10.3389/fpsyg.2017.02314
HapMap (2005) A haplotype map of the human genome. Nature 437:1299–1320
Hardy GH (1908) Mendelian proportions in a mixed population. Science 28:49–50
Hillis DM, Moritz C, Porter CA, Baker RJ (1991) Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251:308–310. https://doi.org/10.1126/science.1987647
Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079. https://doi.org/10.1126/science.1105436
Huelsenbeck JP, Hillis DM (1993) Success of phylogenetic methods in the four-taxon case. Syst Biol 42:247–264. https://doi.org/10.1093/sysbio/42.3.247
Hurst LD (2009) Evolutionary genomics and the reach of selection. J Biol 8:12. https://doi.org/10.1186/jbiol113
Imai A, Nakaya A, Fahiminiya S, Tetreault M, Majewski J, Sakata Y, Takashima S, Lathrop M, Ott J (2015) Beyond homozygosity mapping: family-control analysis based on hamming distance for prioritizing variants in exome sequencing. Sci Rep 5:12028. https://doi.org/10.1038/srep12028
Article PubMed PubMed Central Google Scholar
Imai A, Kohda M, Nakaya A, Sakata Y, Murayama K, Ohtake A, Lathrop M, Okazaki Y, Ott J (2016) HDR: a statistical two-step approach successfully identifies disease genes in autosomal recessive families. J Hum Genet 61:959–963. https://doi.org/10.1038/jhg.2016.85
Inoue I, Nakajima T, Williams CS, Quackenbush J, Puryear R, Powers M, Cheng T, Ludwig EH, Sharma AM, Hata A, Jeunemaitre X, Lalouel JM (1997) A nucleotide substitution in the promoter of human angiotensinogen is associated with essential hypertension and affects basal transcription in vitro. J Clin Invest 99:1786–1797. https://doi.org/10.1172/JCI119343
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, Chow ED, Kanterakis E, Gao H, Kia A, Batzoglou S, Sanders SJ, Farh KK (2019) Predicting splicing from primary sequence with deep learning. Cell 176(535–548):e24. https://doi.org/10.1016/j.cell.2018.12.015
Jeffreys AJ, Wilson V, Thein SL (1985) Hypervariable 'minisatellite' regions in human DNA. Nature 314:67–73. https://doi.org/10.1038/314067a0
Kimura M (1960) Optimum mutation rate and degree of dominance as determined by the principle of minimum genetic load. J Genet 57:21–34
Kimura M (1964) Diffusion models in population genetics. J Appl Probab 1:177–232
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Kimura M (1969) The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61:893–903
Kimura M (1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267:275–276
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Kimura M, Ohta T (1973) The age of a neutral mutant persisting in a finite population. Genetics 75:199–212
King JL, Jukes TH (1969) Non-Darwinian evolution. Science 164:788–798
Kingman JF (2000) Origins of the coalescent. 1974–1982. Genetics 156:1461–1463
Kondrashov AS (1995) Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J Theor Biol 175:583–594. https://doi.org/10.1006/jtbi.1995.0167
Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A (2008) Patterns of positive selection in six Mammalian genomes. PLoS Genet 4:e1000144. https://doi.org/10.1371/journal.pgen.1000144
Kumar S, Tamura K, Nei M (1994) MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191
Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570
Lewontin RC (1972) The apportionment of human diversity. In: Dobzhansky T, Hecht MK, Steere WC (eds) Evolutionary biology, vol 6. Appleton-Century-Crofts, New York, pp 381–398
Lewontin RC, Hubby JL (1966) A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54:595–609
Lipson M, Ribot I, Mallick S, Rohland N, Olalde I, Adamski N, Broomandkhoshbacht N, Lawson AM, Lopez S, Oppenheimer J, Stewardson K, Asombang RN, Bocherens H, Bradman N, Culleton BJ, Cornelissen E, Crevecoeur I, de Maret P, Fomine FLM, Lavachery P, Mindzie CM, Orban R, Sawchuk E, Semal P, Thomas MG, Van Neer W, Veeramah KR, Kennett DJ, Patterson N, Hellenthal G, Lalueza-Fox C, MacEachern S, Prendergast ME, Reich D (2020) Ancient West African foragers in the context of African population history. Nature 577:665–670. https://doi.org/10.1038/s41586-020-1929-1
Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda A, Kikuta K, Takagi Y, Harada KH, Fujiyama A, Herzig R, Krischek B, Zou L, Kim JE, Kitakaze M, Miyamoto S, Nagata K, Hashimoto N, Koizumi A (2011) Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 6:e22542. https://doi.org/10.1371/journal.pone.0022542
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature 461:747–753
Mendel GJ (1866) Versuche über Pflanzen-Hybriden. Verh Naturforsch Ver Brünn 4:3–47
Miyata T, Hayashida H (1981) Extraordinarily high evolutionary rate of pseudogenes: evidence for the presence of selective pressure against changes between synonymous codons. Proc Natl Acad Sci USA 78:5739–5743. https://doi.org/10.1073/pnas.78.9.5739
Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Y, Yamaguchi-Kabata Y, Yokozawa J, Danjoh I, Saito S, Sato Y, Mimori T, Tsuda K, Saito R, Pan X, Nishikawa S, Ito S, Kuroki Y, Tanabe O, Fuse N, Kuriyama S, Kiyomoto H, Hozawa A, Minegishi N, Douglas Engel J, Kinoshita K, Kure S, Yaegashi N, To MJRPP, Yamamoto M (2015) Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun 6:8018. https://doi.org/10.1038/ncomms9018
Nei M (2005) Selectionism and neutralism in molecular evolution. Mol Biol Evol 22:2318–2342. https://doi.org/10.1093/molbev/msi242
Nei M, Roychoudhury AK (1972) Gene differences between Caucasian, Negro, and Japanese populations. Science 177:434–436
Nei M, Roychoudhury AK (1974) Genic variation within and between the three major races of man, Caucasoids, Negroids, and Mongoloids. Am J Hum Genet 26:421–443
Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zollner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V (2012) An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337:100–104. https://doi.org/10.1126/science.1217876
Neyman J (1971) Molecular studies of evolution: a source of novel statistical problems. In: Gupta SS, Yackel J (eds) Statistical decision theory and related topics. Academic Press, New York, pp 1–27
Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andres AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, Indap A, Bustamante CD, Clark AG (2009) Darwinian and demographic forces affecting human protein coding genes. Genome Res 19:838–849. https://doi.org/10.1101/gr.088336.108
Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E (2017) Tracing the peopling of the world through genomics. Nature 541:302–310. https://doi.org/10.1038/nature21347
Ohno S (1970) Evolution by gene duplication. Springer, New York
Ohno S (1972) So much "junk" DNA in our genome. Brookhaven Symp Biol 23:366–370
Ohta T (1973) Slightly deleterious mutant substitutions in evolution. Nature 246:96–98
Ohta T (1992) The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23:263–286
Ohta T (2002) Near-neutrality in evolution of genes and gene regulation. Proc Natl Acad Sci USA 99:16134–16137. https://doi.org/10.1073/pnas.252626899
Pattison JE (2016) An attempt to integrate previous localized estimates of human inbreeding for the whole of Britain. Hum Biol 88:264–274
PubMed Google Scholar
Provine WB (1971) The origins of theoretical population genetics. University of Chicago Press, Chicago
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sella G, Barton NH (2019) Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu Rev Genomics Hum Genet 20:461–493. https://doi.org/10.1146/annurev-genom-083115-022316
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Sundaram L, Gao H, Padigepati SR, McRae JF, Li Y, Kosmicki JA, Fritzilas N, Hakenberg J, Dutta A, Shon J, Xu J, Batzoglou S, Li X, Farh KK (2018) Predicting the clinical impact of human mutation with deep neural networks. Nat Genet 50:1161–1170. https://doi.org/10.1038/s41588-018-0167-z
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Takahata N, Nei M (1985) Gene genealogy and variance of interpopulational nucleotide differences. Genetics 110:325–344
Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J (2020) On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7:1012–1023. https://doi.org/10.1093/nsr/nwaa036
Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, Project NES (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337:64–69. https://doi.org/10.1126/science.1219240
Weinberg W (1908) Über den Nachweis der Vererbung beim Menschen. Jahreshefte des Vereins für vaterländische Naturkunde in Württemberg 64:369–382
Williams GC, Nesse RM (1991) The dawn of Darwinian medicine. Q Rev Biol 66:1–22. https://doi.org/10.1086/417048
Wright S (1938) Size of population and breeding structure in relation to evolution. Science 87:430–431
Yang J, Zeng J, Goddard ME, Wray NR, Visscher PM (2017) Concepts, estimation and interpretation of SNP-based heritability. Nat Genet 49:1304–1310. https://doi.org/10.1038/ng.3941
Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A (2019) A primer on deep learning in genomics. Nat Genet 51:12–18. https://doi.org/10.1038/s41588-018-0295-5
Download references
Acknowledgements
Helpful comments by Prof. Joseph Felsenstein on an earlier version of this manuscript are gratefully acknowledged. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI, Grant numbers JP20K08497 and JP18K15863 (A. O.), and Grant number 19K09408 (S. Y.).
Author information
Authors and affiliations.
Intractable Disease Research Center, Juntendo University, Tokyo, Japan
Atsuko Okazaki
Laboratory of Statistical Genetics, Rockefeller University, 1230 York Avenue, New York, NY, 10065, USA
Atsuko Okazaki & Jurg Ott
Department of Molecular Pharmacology, National Cerebral and Cardiovascular Center, Osaka, Japan
Satoru Yamazaki
Division of the Human Genetics, National Institute of Genetics, Shizuoka, Japan
Ituro Inoue
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Jurg Ott .
Ethics declarations
Conflict of interest.
The authors declare no conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Okazaki, A., Yamazaki, S., Inoue, I. et al. Population genetics: past, present, and future. Hum Genet 140 , 231–240 (2021). https://doi.org/10.1007/s00439-020-02208-5
Download citation
Received : 12 May 2020
Accepted : 14 July 2020
Published : 18 July 2020
Issue Date : February 2021
DOI : https://doi.org/10.1007/s00439-020-02208-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Find a journal
- Publish with us
- Track your research
- Support Our Work
The Smithsonian Institution's Human Origins Program
- Introduction to Human Evolution

Human evolution
Human evolution is the lengthy process of change by which people originated from apelike ancestors. Scientific evidence shows that the physical and behavioral traits shared by all people originated from apelike ancestors and evolved over a period of approximately six million years.
One of the earliest defining human traits, bipedalism -- the ability to walk on two legs -- evolved over 4 million years ago. Other important human characteristics -- such as a large and complex brain, the ability to make and use tools, and the capacity for language -- developed more recently. Many advanced traits -- including complex symbolic expression, art, and elaborate cultural diversity -- emerged mainly during the past 100,000 years.
Humans are primates. Physical and genetic similarities show that the modern human species, Homo sapiens , has a very close relationship to another group of primate species, the apes. Humans and the great apes (large apes) of Africa -- chimpanzees (including bonobos, or so-called “pygmy chimpanzees”) and gorillas -- share a common ancestor that lived between 8 and 6 million years ago. Humans first evolved in Africa, and much of human evolution occurred on that continent. The fossils of early humans who lived between 6 and 2 million years ago come entirely from Africa.
Most scientists currently recognize some 15 to 20 different species of early humans. Scientists do not all agree, however, about how these species are related or which ones simply died out. Many early human species -- certainly the majority of them – left no living descendants. Scientists also debate over how to identify and classify particular species of early humans, and about what factors influenced the evolution and extinction of each species.
Early humans first migrated out of Africa into Asia probably between 2 million and 1.8 million years ago. They entered Europe somewhat later, between 1.5 million and 1 million years. Species of modern humans populated many parts of the world much later. For instance, people first came to Australia probably within the past 60,000 years and to the Americas within the past 30,000 years or so. The beginnings of agriculture and the rise of the first civilizations occurred within the past 12,000 years.
Paleoanthropology
Paleoanthropology is the scientific study of human evolution. Paleoanthropology is a subfield of anthropology, the study of human culture, society, and biology. The field involves an understanding of the similarities and differences between humans and other species in their genes, body form, physiology, and behavior. Paleoanthropologists search for the roots of human physical traits and behavior. They seek to discover how evolution has shaped the potentials, tendencies, and limitations of all people. For many people, paleoanthropology is an exciting scientific field because it investigates the origin, over millions of years, of the universal and defining traits of our species. However, some people find the concept of human evolution troubling because it can seem not to fit with religious and other traditional beliefs about how people, other living things, and the world came to be. Nevertheless, many people have come to reconcile their beliefs with the scientific evidence.
Early human fossils and archeological remains offer the most important clues about this ancient past. These remains include bones, tools and any other evidence (such as footprints, evidence of hearths, or butchery marks on animal bones) left by earlier people. Usually, the remains were buried and preserved naturally. They are then found either on the surface (exposed by rain, rivers, and wind erosion) or by digging in the ground. By studying fossilized bones, scientists learn about the physical appearance of earlier humans and how it changed. Bone size, shape, and markings left by muscles tell us how those predecessors moved around, held tools, and how the size of their brains changed over a long time. Archeological evidence refers to the things earlier people made and the places where scientists find them. By studying this type of evidence, archeologists can understand how early humans made and used tools and lived in their environments.
The process of evolution
The process of evolution involves a series of natural changes that cause species (populations of different organisms) to arise, adapt to the environment, and become extinct. All species or organisms have originated through the process of biological evolution. In animals that reproduce sexually, including humans, the term species refers to a group whose adult members regularly interbreed, resulting in fertile offspring -- that is, offspring themselves capable of reproducing. Scientists classify each species with a unique, two-part scientific name. In this system, modern humans are classified as Homo sapiens .
Evolution occurs when there is change in the genetic material -- the chemical molecule, DNA -- which is inherited from the parents, and especially in the proportions of different genes in a population. Genes represent the segments of DNA that provide the chemical code for producing proteins. Information contained in the DNA can change by a process known as mutation. The way particular genes are expressed – that is, how they influence the body or behavior of an organism -- can also change. Genes affect how the body and behavior of an organism develop during its life, and this is why genetically inherited characteristics can influence the likelihood of an organism’s survival and reproduction.
Evolution does not change any single individual. Instead, it changes the inherited means of growth and development that typify a population (a group of individuals of the same species living in a particular habitat). Parents pass adaptive genetic changes to their offspring, and ultimately these changes become common throughout a population. As a result, the offspring inherit those genetic characteristics that enhance their chances of survival and ability to give birth, which may work well until the environment changes. Over time, genetic change can alter a species' overall way of life, such as what it eats, how it grows, and where it can live. Human evolution took place as new genetic variations in early ancestor populations favored new abilities to adapt to environmental change and so altered the human way of life.
Dr. Rick Potts provides a video short introduction to some of the evidence for human evolution, in the form of fossils and artifacts.
- Climate Effects on Human Evolution
- Survival of the Adaptable
- Human Evolution Timeline Interactive
- 2011 Olorgesailie Dispatches
- 2004 Olorgesailie Dispatches
- 1999 Olorgesailie Dispatches
- Olorgesailie Drilling Project
- Kanam, Kenya
- Kanjera, Kenya
- Ol Pejeta, Kenya
- Olorgesailie, Kenya
- Evolution of Human Innovation
- Adventures in the Rift Valley: Interactive
- 'Hobbits' on Flores, Indonesia
- Earliest Humans in China
- Bose, China
- Anthropocene: The Age of Humans
- Fossil Forensics: Interactive
- What's Hot in Human Origins?
- Instructions
- Carnivore Dentition
- Ungulate Dentition
- Primate Behavior
- Footprints from Koobi Fora, Kenya
- Laetoli Footprint Trails
- Footprints from Engare Sero, Tanzania
- Hammerstone from Majuangou, China
- Handaxe and Tektites from Bose, China
- Handaxe from Europe
- Handaxe from India
- Oldowan Tools from Lokalalei, Kenya
- Olduvai Chopper
- Stone Tools from Majuangou, China
- Middle Stone Age Tools
- Burin from Laugerie Haute & Basse, Dordogne, France
- La Madeleine, Dordogne, France
- Butchered Animal Bones from Gona, Ethiopia
- Katanda Bone Harpoon Point
- Oldest Wooden Spear
- Punctured Horse Shoulder Blade
- Stone Sickle Blades
- Projectile Point
- Oldest Pottery
- Pottery Fragment
- Fire-Altered Stone Tools
- Terra Amata Shelter
- Qafzeh: Oldest Intentional Burial
- Assyrian Cylinder Seal
- Blombos Ocher Plaque
- Ishango Bone
- Bone and Ivory Needles
- Carved Ivory Running Lion
- Female torso in ivory
- Ivory Horse Figurine
- Ivory Horse Sculpture
- Lady of Brassempouy
- Lion-Man Figurine
- Willendorf Venus
- Ancient Shell Beads
- Carved Bone Disc
- Cro-Magnon Shell Bead Necklace
- Oldest Known Shell Beads
- Ancient Flute
- Ancient Pigments
- Apollo 11 Plaque
- Carved antler baton with horses
- Geometric incised bone rectangle
- Tata Plaque
- Mystery Skull Interactive
- Shanidar 3 - Neanderthal Skeleton
- One Species, Living Worldwide
- Human Skin Color Variation
- Ancient DNA and Neanderthals
- Human Family Tree
- Swartkrans, South Africa
- Shanidar, Iraq
- Walking Upright
- Tools & Food
- Social Life
- Language & Symbols
- Humans Change the World
- Nuts and bolts classification: Arbitrary or not? (Grades 6-8)
- Comparison of Human and Chimp Chromosomes (Grades 9-12)
- Hominid Cranial Comparison: The "Skulls" Lab (Grades 9-12)
- Investigating Common Descent: Formulating Explanations and Models (Grades 9-12)
- Fossil and Migration Patterns in Early Hominids (Grades 9-12)
- For College Students
- Why do we get goose bumps?
- Chickens, chimpanzees, and you - what do they have in common?
- Grandparents are unique to humans
- How strong are we?
- Humans are handy!
- Humans: the running ape
- Our big hungry brain!
- Our eyes say it!
- The early human tool kit
- The short-haired human!
- The “Nutcracker”
- What can lice tell us about human evolution?
- What does gut got to do with it?
- Why do paleoanthropologists love Lucy?
- Why do we have wisdom teeth?
- Human Origins Glossary
- Teaching Evolution through Human Examples
- Frequently Asked Questions
- Recommended Books
- Exhibit Floorplan Interactive
- Print Floorplan PDF
- Reconstructions of Early Humans
- Chesterfield County Public Library
- Orange County Library
- Andover Public Library
- Ephrata Public Library
- Oelwein Public Library
- Cedar City Public Library
- Milpitas Library
- Spokane County Library
- Cottage Grove Public Library
- Pueblo City-County Library
- Springfield-Greene County Library
- Peoria Public Library
- Orion Township Public Library
- Skokie Public Library
- Wyckoff Free Public Library
- Tompkins County Public Library
- Otis Library
- Fletcher Free Library
- Bangor Public Library
- Human Origins Do it Yourself Exhibit
- Exhibit Field Trip Guide
- Acknowledgments
- Human Origins Program Team
- Connie Bertka
- Betty Holley
- Nancy Howell
- Lee Meadows
- Jamie L. Jensen
- David Orenstein
- Michael Tenneson
- Leonisa Ardizzone
- David Haberman
- Fred Edwords (Emeritus)
- Elliot Dorff (Emeritus)
- Francisca Cho (Emeritus)
- Peter F. Ryan (Emeritus)
- Mustansir Mir (Emeritus)
- Randy Isaac (Emeritus)
- Mary Evelyn Tucker (Emeritus)
- Wentzel van Huyssteen (Emeritus)
- Joe Watkins (Emeritus)
- Tom Weinandy (Emeritus)
- Members Thoughts on Science, Religion & Human Origins (video)
- Science, Religion, Evolution and Creationism: Primer
- The Evolution of Religious Belief: Seeking Deep Evolutionary Roots
- Laboring for Science, Laboring for Souls: Obstacles and Approaches to Teaching and Learning Evolution in the Southeastern United States
- Public Event : Religious Audiences and the Topic of Evolution: Lessons from the Classroom (video)
- Evolution and the Anthropocene: Science, Religion, and the Human Future
- Imagining the Human Future: Ethics for the Anthropocene
- Human Evolution and Religion: Questions and Conversations from the Hall of Human Origins
- I Came from Where? Approaching the Science of Human Origins from Religious Perspectives
- Religious Perspectives on the Science of Human Origins
- Submit Your Response to "What Does It Mean To Be Human?"
- Volunteer Opportunities
- Submit Question
- "Shaping Humanity: How Science, Art, and Imagination Help Us Understand Our Origins" (book by John Gurche)
- What Does It Mean To Be Human? (book by Richard Potts and Chris Sloan)
- Bronze Statues
- Reconstructed Faces
- Open access
- Published: 29 October 2018
RETRACTED ARTICLE: A brief history of human evolution: challenging Darwin’s claim
- Sarah Umer ORCID: orcid.org/0000-0002-2725-1016 1
International Journal of Anthropology and Ethnology volume 2 , Article number: 6 ( 2018 ) Cite this article
44k Accesses
3 Citations
101 Altmetric
Metrics details
This article was retracted on 19 February 2019
This article has been updated
There is a consensus among evolutionists today that man first appeared in Africa approximately four million years ago. Others counter this theory saying, “... when shall we speak of man as man”? The timeline they give is approximately one million years and to fully understand one million years is still a difficult task.
However, another even better way to understand time and man is to study it in terms of generations. So, keeping in mind that primitive people married and had children early, twenty years will make an average generation. According to this there would be 50,000 generations in a million years. Keeping this in mind if we calculate generations we find that 250 generations back take us to the time when written history began. While, another 250 generations back would take us to the time (10,000 years ago), when cultivation began, and man started settled life. Now we are left with 49,500 generations of men, plus a time span of 990,000 years. Keeping these statistics in mind let us ask the question once more, when should we speak of man as man?
Therefore, this paper attempts not only to understand the timeframe “when we can really call Man? – Man” in light of the so-called history of human evolution but also to understand that if the specie roaming the earth for a million years was truly man’s ancestor, as is claimed by Charles Darwin. Then what took man’s ancestor so long to show signs of development that we only witness in the last 12000 years.
Moreover, while keeping man’s progress under consideration of the last 12000 years, it will further shed light on why there are serious reservations about Charles Darwin theory of human evolution. As many scientists, evolutionists, archeologist and different religious scriptures strongly claim that man came to the earth fully developed and did not evolve from a lesser specie.
Introduction
Prehistory simply means the time before written history began. More than 99% of man’s story is prehistory. The consensus by the historians is that man is about 1 million years old, but he did not write anything, until 5000 years ago. Although, prehistoric man did not leave us any written records he unintentionally left us information on his way of life, which is interpreted by different kinds of scientists. These scientists are specialist in physical anthropologists, human paleontologists, archaeologist and other fields. It is their job to find out what happened before written history began (Braidwood 1964a , b ). Some historians believe that it is their duty to bring together various facts given by these scientists and put them together in such a way as to reach objective conclusions. E. H. Carr has two ideas on what an objective historian is: one is that he “... has the capacity to rise above the limited vision of his own situation in society and in history – a capacity which... is partly dependent on his capacity to recognize the extent of his involvement in that situation, to recognize, that is to say, the impossibility of total objectivity”, and his other idea is “...simply that he gets his facts right, but rather that he chooses the right facts, or, in other words, that he applies the right standard of significance” (Marwick 1970 ).
Man has been blessed with the best attributes and qualities among all living species. It is one, specie that can control all other species with its intelligence and wisdom. It is the one specie that has the ability to reason and think rationally. C. M. Bowra says, “based on the humanistic faith that man is worth studying for his own sake”, that “Human beings are the element in our environment which is of most consequence to every child of man” (Grant 1965 , 1952 ). Therefore we should not be a surprised when we see many theories and counter theories with regard to the dates of the origin, the evolution or the different phases of this development.
An enormous span of human history needs to be covered in order to have answers to these questions. Robert J. Braidwood, in the preface to his book, Prehistoric Men , states,
“New discoveries and new techniques for the interpretation of the evidence of mankind’s past appear almost daily. The newer finds and techniques necessitate reconsideration of older evidence. Slowly but surely we move toward fuller understandings of those beings whose history holds the greatest fascination for all of mankind-men themselves.”
Whatever, material I have put together in this paper and whatever hypothesis, assumptions or conclusions, I shall make will be based on the work done up until this time. What the future discoveries behold is still unknown and mysterious to all of us.
There are many different ways and views about how to understand prehistory and history. Pre-historians have given two main ways to accomplish this task. One is either to go down the ladder of time, whereas the other is to climb up the ladder. I will follow the first path for this paper.
There is a general consensus among evolutionists today that man first appeared in Africa approximately 4 million years ago. Others counter this theory saying, “... when shall we speak of man as man”? The timeline they give is approximately 1 million years. Even to fully understand 1 million years is very difficult task. For example, if we compare this whole time period to 1 day, it would be something like this:
“The present time is midnight, and Jesus was born just two minutes and fifty seconds ago. Earliest history began about seven minutes ago. Everything before 11:53 P.M. was in prehistoric time” (Braidwood 1964a , 1967b : 10–11).
Another, even better way to understand time is to study it in terms of generations. So, keeping in mind that primitive people married and had children early, 20 years will make an average generation. According to this there would be 50,000 generations in a million years. Keeping this in mind if we calculate generations we find that 250 generations back take us to the time when written history began,
“...David was king of Israel less than 150 generations ago, Julius Caesar was alive just 100 generations ago, Columbus was there 25 generations ago and the United States is just 10 generations old. So, the current scenario is that there were 49,750 generations of men before written history began!” (Braidwood 1964a , 1967b )
And another 250 generations back would take us to the time (10,000 years ago), when cultivation began and man started settled life. Now we are left with 49,500 generations of men, plus a time span of 990,000 years. Keeping these statistics in mind let us ask the question once more, when should we speak of man as man? Can the human mind accept this fact that for such a long period of time and for so many generations, man did not make any effort to change either his surrounding or his life style? Why was he only a hunter and a gatherer for thousands and thousands of years? Why did it take him such a long time to change himself and his way of life? Or would it not be wrong to say here that it was not really man who was roaming the earth at that time, but some other species, with fewer qualities than what man has been blessed with. Some human evolutionists have tried to call these species man’s ancestors and they believe that man eventually evolved out of these species, whereas others have refuted this concept. According to them, man did not evolve out of any other species: man was born a man with the best of qualities among all living species.
Results and discussion
Let us briefly review human evolution, in order to understand this concept and to try to find answers to questions that are still confusing us today. Let us begin with the evolution of animals. Zoologists have classified the members of the animal kingdom according to their differences and similarities. We humans fall under this kingdom because we move and eat with our mouth; we are vertebrates because of our backbone, and we are mammals because we are warm-blooded and we breast-feed our offspring. We are primates because we have grasping hands, flexible limbs, and a highly developed sense of vision. We are also members of the family Hominoidea , the taxonomic group which includes both humans and apes, because of the absence of a tail, swinging arms, and the shape of our teeth. The term hominoid refers to all present and past apes and humans, while hominid refers specifically to present and past humans (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ). Hominids are known as walking creatures with comparatively large brains; humans today are the sole living representation of this group. According to some evolutionists, fossil bones and genetic studies indicate that the hominids shared an ancestor with the great African ape. It is at this point that the record becomes more complex, when the study of primate evolution turns into the study of hominid evolution. According to Richard Klein of Stanford University, determining the genus and species of the fossil bones of early humans is a very difficult task. The fragmentary pieces of the early fossil finds represent only a few hundred separate individuals. Determining the age of the fossils is also very difficult. He says paleo-anthropology is more like a court of law than a physics laboratory, where we reassess and even redraw the whole family tree on finding a new fragment (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ).
Although we humans differ considerably from apes, genetically we are closest to them of all hominoids. As far as our genetic composition is concerned we share 98.4% of our genetic material with chimpanzees, while gorillas are only 2.3% different from us genetically. These statistics show great similarities and suggests that the last ancestor shared by the great ape and human lines was probably a chimpanzee-like creature. Thus, changes in genetic regulatory mechanism play an important role in the evolution of different lineages.
Geography had quite an important influence in shaping the development of humanity. It was in the middle of the Tertiary period (5–25 million of years ago) when the climate was much warmer and wetter than what it is today, and tropical forests grew across much of Africa, Europe and Asia that an increase in the variety of mammals occurred. Many species of apes lived in these forests, including one that is considered by some to be the ancestor of modern humans.
Then sometime around 5 million years ago, towards the end of the Tertiary time, the global temperatures began to cool, ice caps formed at the poles, and the climate grew drier. As a result, the area of the tropical forests grew smaller, giving way to expanses of open woodland and grasslands. These developments did not occur all at once but evolved slowly. In East Africa the hominoids groups were trapped in shrinking patches of forest. Before this they had lived in the trees and moved on four feet when travelling around the forest bed. Now in order to cross wide stretches of open land quickly, some hominoids began walking on two feet, like modern humans. It was under these conditions that the hominids split from this ape or the hominoid group. It is believed that this whole process of hominization began in Africa, which is the only continent where fossils of early hominids dating back to four - 5 million years are found today. The distinction was made due to their new mode of locomotion, known as “bipedalism” (walking on two feet rather than on four). These early hominids are called australopithecines: specialist believe in addition to walking upright with two feet, they had comparatively larger brains and they depended on tools for their survival (Bahn 2002a , b , c , d , e ).
While studying the fossils we also need to study how these beings modified objects and landscapes, thus creating an archaeological record. It is important to study both, as it helps us to understand how human beings evolved into what they are today. Some of the best evidence comes from sites at Hadar, Swartkrans, Olduvai, Laetoli and Koobi Fora. Some new fossil finds from Ethiopia, Kenya and Tanzania have pushed back the age of the earliest known hominids and have modified our understanding of their appearance and behavior (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ).
The oldest known Australopithecus species is A. Anamensis , which dates approximately 4 million years ago. The remaining early hominid fossils have been assigned to the species A. Afarensis , which includes the famous Lucy from Hadar (see Fig. 1 - A. Afarensis, Lucy from Hadar, Ethiopia), Ethiopia. Other direct evidence of hominid bipedalism is a fossilized trail of footprints some 3.6 million years old, found in eastern Africa at Laetoli, Tanzania (see Fig. 2 - Replica of the fossilized trail of footprints some 3.6 million years old, found in eastern Africa at Laetoli, Tanzania are exhibited in the National Museum of Nature and Science, Tokyo, Japan). Another evolutionary trend that occurred during this time was the change in dental pattern. Fossils of hominids dating back to three to 2 million years from southern and eastern African sites have small front teeth and large cheek teeth. Some form of gracile australopithecine is thought to have evolved into the first members of the genus homo about 2 million years ago, known as Homo habilis (which literally means handyman). Anthropologists continue to disagree about what caused this complete transition from ape to humans. Whereas many paleontologists believe that more than one species belonging to the genus homo may have co-existed in eastern Africa during the early Pleistocene, along with the robust australopithecines (species with massive teeth and jaws). It is also during this time that we find the oldest undisputed stone tools from Ethiopia, classified under the Oldowan tradition. By 1.8 million years ago the early homos had either disappeared or they evolved into H. erectus (upright man), the first member of the genus homo to spread out of Africa into Asia and Europe (see Fig. 3 - H. erectus (upright man), the first member of the genus homo to spread out of Africa into Asia and Europe.). They were taller than modern humans, but were very much like us in other respects, though their brains were still much smaller. It is assumed that they were capable of using fire and speech to a certain extent, and went, to far areas like Central Asia, Southwest Asia, South Asia, Europe and China. The Acheulean stone-tool tradition is associated with them. Archaeologists traditionally assign the Oldowan and Acheulean traditions to a single period known as the Lower Paleolithic in Europe and the Early Stone Age in Africa. But in recent years archaeologists have concluded that it is quite misleading to associate any Stone Age traditions uniquely to a single hominid species. Traditions can vary due to the availability of particular resources in different areas. Keeping all the above details in mind, paleoanthropologists consider a logical link between H. erectus , the more primitive hominids and our own species. They believe that the Acheulean stone – tool making ability was a determinant factor in the migration of early hominids out of Africa into new environments. The earliest handaxes found from outside of Africa are in Ubeidiya, Israel. While the punctuationists and gradualists continue to debate about the fate of H. erectus and the origin of the H. sapiens , up to this time indicates that our own species issued from H. erectus (Schultz and Lavenda 1998a , b , c , d ).
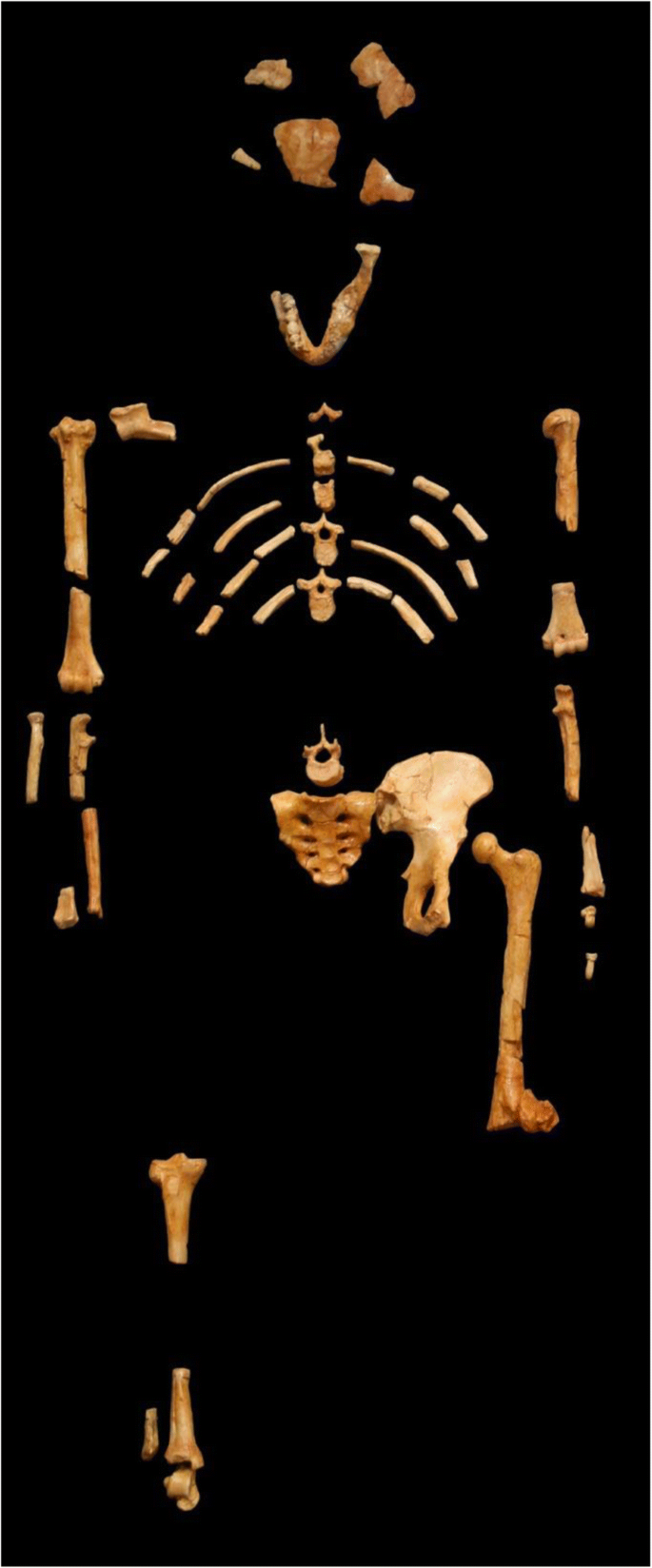
A. Afarensis , Lucy from Hadar, Ethiopia ( https://en.wikipedia.org/wiki/Lucy_(Australopithecus )

Replica of the fossilized trail of footprints some 3.6 million years old, found in eastern Africa at Laetoli, Tanzania are exhibited in the National Museum of Nature and Science, Tokyo, Japan

H. erectus (upright man), the first member of the genus homo to spread out of Africa into Asia and Europe. ( http://animals.wikia.com/wiki/Homo_erectus )
Towards the end of the Tertiary Period the global climate had started to cool down and this continued into the next Quaternary Period, also known as the Ice Age (see Fig. 4 - Map of the Last Ice Age.). In spite of its name the climate was not cold all the time: there were frequent warm intervals interglacial periods, separate from the cold, dry glacial periods. As these changes were quite rapid, the animals and plants sometimes found it difficult and sometimes impossible to adapt and to survive under the new climatic changes. H. erectus responded to these changes by developing a bigger brain. This meant that greater intelligence was available now for problem solving. One million years ago the brain of H. erectus was approximately three - quarters the size of the modern human brain (Haywood 1997 , 1989 n.d. ,). But recent studies have shown that stone tools found in association with animal bones were not used for slaughtering animals but for cleaning the skins and cutting up the meat. Plants probably formed a large part of their diet. No ornaments or art work is found and neither is there any evidence of them burying their dead (Bahn 2002a , b , c , d , e ). So the question remains, was H. erectus man’s ancestor?
Map of the Last Ice Age. ( http://www.kerbtier.de/Pages/Themenseiten/enPhylogenie.html )
It was sometime between 500,000 and 200,000 years ago that the fossils of H. erectus start to disappear from the fossil records and were replaced by fossils that show a mosaic of features found in H. erectus and H. sapiens (literally wise men). Though these fossils differ considerably from one another they collectively are known as archaic H. sapiens . They are poorly dated and paleoanthropologists find them difficult to classify and to relate specifically to H. sapiens . There are two major interpretations of the evidence of these fossil records. Punctuationists favor the “replacement model”: according to them H. erectus was a single, long- lived, geographically dispersed species, and had only one sub-population most probably located in Africa around 200,000 to 130,000 years ago that underwent evolutionary changes there to produce H. sapiens . Their descendants then later migrated to other regions. Whereas, the gradualists favor the “regional continuity model”. According to it H. erectus eventually evolved into H. sapiens gradually throughout the entire regions, retaining regionally distinct physical characteristics (Schultz and Lavenda 1998a , b , c , d ). However, archaeologists have found no record to back the claim that either H. erectus or archaic H. sapiens was truly our ancestor. There is no evidence of personal ornamentation - jewelry, beads, or any kinds of art form, exists, nor are there paintings, sculptures or engravings that show that use of more than basic instincts. The ability to think and reason is still missing (Ingold 1994a , b , c ). Therefore, whether they were really our ancestor, might be questioned.
In Europe a specie known as Neanderthals flourished between 130,000 and 35,000 years ago, they are considered by some to be the descendant of archaic H. sapiens . A large collection of fossils remains, tells us that they were shorter and more robust than modern H. sapiens with bulbous noses that helped them to survive cold conditions by reducing heat loss. But during the 1980s, new evidence revealed that Neanderthals appeared in Europe and western Asia at the same time anatomically modern H. sapiens appeared in Africa. These dates and the new evidence have made paleoanthropologists revise their traditional understanding of the relationship between Neanderthals and modern peoples. The archaic H. sapiens could no longer be considered the ancestors of the Neanderthals as data found in Israel suggests that the Neanderthals and the moderns lived side by side in south-western Asia for at least 45,000 years without losing their anatomical distinctiveness. Even more puzzling is the fact that both were using tools made in the same way. Mousterian experts disagree whether Neanderthals created a religion and whether hunting was important to them, but there is compelling fossil evidence from many sites and regions that they buried their dead and looked after their sick and old. Moreover, it seems probable, given the scant evidence for any form of art or ornamentation, that they did not make use of symbols, which is a critical element in the development of human language. Another thing that concerns us today is whether the Neanderthals and the moderns interbred, and whether the modern human populations today contain any Neanderthal genes. This situation creates a dispute among historians today (Schultz and Lavenda 1998a , b , c , d ). There are many questions that come to mind. Why did the Neanderthals look after their sick and old? Why did they start to bury their dead? Why were they using tools? Some believe that all these actions can be related to basic instincts. Tool use is not a distinctive characteristic of humans but; animals too may use or even make simple tools; however, using tools to make other tools does distinguish humans from animals. For example, the sea otter wields a rock to break open the shell of an abalone. And the anthropologist Jane Goodall has observed chimpanzees using a variety of tools in their daily life: thrashing about with branches for display, using clubs and missiles for defense, selecting a twig and stripping its bark to probe the nests of termites and attract them to the stick, then to be eaten by the wise chimp. West African chimps even use stone and wooden hammers to crack and open nutshells (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ).
So, the question becomes was the Neanderthal man’s ancestor? Because when it came to something like creating symbols for speech they were unable to do so, since it required more brain capacity, reasoning and intelligence that they unfortunately lacked. The question whether the Neanderthals and modern humans interbred was recently addressed by paleoanthropologists who claim, that there was no interbreeding between the two. Mitochondrial DNA studies suggest that all humans living today are part of a relatively homogeneous population that originated in Africa within the last few hundred thousand years. In 1997, genetic researchers extracted and decoded a mitochondrial DNA fragment for the original Neander Valley specimen. The analysis revealed significant differences in its DNA from all living humans, suggesting that there was an ancient split between the two lineages, perhaps more than 500,000 years ago. Although, Neanderthals did not disappear from Western Europe until 30,000 years ago, possibly later, it is a common belief that modern H. sapiens may have forced them into extinction (Bahn 2002a , b , c , d , e ).
Now let’s enter the last and the most important phase of human evolution. This phase is further divided into two phases. During the first phase, there is a general consensus among paleoanthropologists today that modern human ( H. sapiens ) evolved in Africa sometime between 100,000 to 150,000 years ago and spread around the globe. Recent studies in genetic evolution also support the view that Africa was the home of the original human population. However, debate continues about the nature of their dispersal. Most believe that a spreading wave of modern humans replaced existing populations of archaic H. sapiens entirely. This process of dispersal was complex and involved multiple movements of people and genes. In the caves of Qafzeh and Skhul in Israel remains of modern humans similar to found in Ethiopia and Tanzania, (150,000–100,000 years ago), and in South Africa (100,000–90,000 years ago) have been found. This is the first evidence that we have of modern humans (if they were) outside of Africa. Though, the fossils of these modern humans are still associated with the archaic stone tool traditions (like those of the Neanderthals ), and like the ones also associated with the race of modern Africans (Bahn 2002a , b , c , d , e ).
According to paleoanthropologists a second phase began roughly around 40,000 years ago, when “a behavioral revolution” took place: whether it was the continuity of the race or whether the race of the: “so called out of Africa H. sapiens ” was totally replaced by the current human race are two questions that are under continuing discussion. But no one can disregard or refute the dramatic changes of many both in anatomy and in behavior, that have taken place over the last 40,000 years when compared with the previous million years. Recent evidence from molecular biology has added support to this picture of rapid and recent change, resulting in the current humans that are not only genetically but also behaviorally and anatomically modern. Evidence also points to an African center for the origin of modern humans. As they moved out of Africa they very soon replaced the variety of other Homo geneses roaming the world (Ingold 1994a , b , c ).
Modern humans besides having many biological differences from other homo species are according to some “closer to the angles”. We possess many attributes that differentiate us from other species. Our large brain and intelligence enables us to think rationally and make decisions rather than to follow basic instincts like other species. We as humans have moved from purely instinctual behavior to reason and thought. We, in a given situation may flee from a fire, but we can also turn back into the same fire to save someone else (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ). It is during this phase, that we see fully developed linguistic and modern technological skills which appear to have developed in the modern man. There is general disagreement as to when this really happened, as the evidence found in this regard is both uncertain and open to doubt. Some believe it was 100,000 years ago, whereas others say it was as recent as 50,000 to 40,000 years ago. However, according to the archaeological record, it is only after 50,000 years that we find abundant examples of art and advanced technology. The first uncontested ritual behavior evidence that we have are the ostrich eggshells beads found from Enkapune ya Muto (Kenya) dating to 46,000 years ago. Beside these archaeologists also witness the appearance of ornaments, engravings, sculptures, and other form of symbolism, which unmistakably confirm the presence of modern human language. But the fossils of this time remain ambiguous, because they lack any anatomical evidence for linguistic abilities. Despite lack of evidence found in the fossils, the archaeological record speaks louder than words. The manipulation of these symbols (ornaments, engravings, etc.) are linked with the fundamental improvements that occurred in technical abilities, which undoubtedly played an important role in the global spread of the modern human species. Their ability to invent new technologies and cope with different environments helped them to colonize the globe at a rapid speed (Bahn 2002a , b , c , d , e ).
So, it would not be wrong to presume at this stage, that it was the abilities to talk, think, reason and communicate that differentiated modern humans from all other creatures before him. Clifford Geertz of Princeton University has described humans as,
“...toolmaking, talking, symbolizing animals: Only they laugh; only they know when they will die; only they disdain to mate with family members; only they contrive those visions of other worlds called art. They have not just mentality but consciousness, not just needs but values, not just fears but conscience, not just a past but a history. Only they have culture.”
According to the famous anthropologist Leslie White, culture is our “extrasomatic” means of survival, it is the nonbiological, nongenetic behavior and sociability that have carried us through the millennia and spread us into diverse environments across the planet. So, in short, culture is a group of ideas and actions that are learned and transmitted from one generation to the next generation. Human culture embodies our experiences and behaviors which are summarized in our language and are transferred to us through our parents and peers. It is as impossible to have human identity without social contact as it is to have biological existence without parents. There is a famous story that Tarzan of the comic book and movie was an ape before he met Jane. It is only culture that enables us to find our place on earth, to create Gods, to anticipate death, to travel to the worlds beyond, and last but not the least to study archaeology, in order to find answers about our past (Price and Feinman 1993 , 1997a , 1997b , c , d , e , f ). It was this cultural development, which was both very rapid and at an alarming speed, that until today evolutionists, biological scientist, paleoanthropologist and many more have been unable to understand. For example, if we can believe the evolutionist man’s ancestor first appeared on earth 4 million years ago and then slowly evolved into Modern Human only around 40,000 years ago. While keeping this in mind we witness, that after 40,000 to 10,000 years age man not only developed new technologies, but he also modified his environment. And only 10,000 years have taken him from bows and arrows to thermonuclear weapons, and the production of the latter has taken only twenty more years (The New Encyclopedia Britannica n.d. ).
Another important fact that seems to refute the claim of the evolutionists today is that, there are no signs of any intermediate forms found in the fossil records. Charles Darwin, who is known as the father of the theory of evolution, as state in his book, The Origins of Species claims,
“If my theory be true, numberless intermediate varieties, linking most closely all of the species of the same group together must assuredly have existed... Consequently, evidence of their former existence could be found only amongst fossil remains.”(Darwin 1964 )
The fossil records today show few intermediate forms; on the other hand, we see fully-formed living species seem to emerge suddenly without any evolutionary transitional form between them. This lack of factual evidence is enough to back their claim that all living species are created separately, and that life appeared on earth all of a sudden and fully-formed. Derek V. Ager, a famous British evolutionist admits this fact by saying;
“The point emerges that if we examine the fossil record in detail, whether at the level of Orders or of Species, we find – over and over again – not gradual evolution, but the sudden explosion of one group at the expense of another.”(Ager 1976 )
The fact that all living species were created separately, suddenly and fully-formed without any evolutionary ancestor is yet again backed by evolutionist biologist Douglas Futuyma, who claimed,
“Creation and evolution, between them, exhaust the possible explanations for the origin of living things. Organisms either appeared on the earth fully developed or they did not. If they did not, they must have developed from pre-existing species by some process of modification. If they did appear in a fully developed state, they must indeed have been created by some omnipotent intelligence.”(Futuyma 1983 )
Fossil records today back this claim that all living species emerged fully developed and in a perfect state on earth.
Let’s counter Charles Darwin’s claim about the ‘origin of man’, that he evolved from some ape-like creatures. The evolutionists who back him claim, that during the 5 million years of man’s evolution, man evolved from one stage of species to another. How these different stages evolved one after the other have already been discussed in detail. So, in short, according to evolutionists who give counter arguments claiming that, the first stage that is, Australopithecus also known as South African ape is nothing but an old ape that has become extinct. Extensive research was carried out by anatomists from both England and America, namely, Lord Solly Zuckerman and Prof. Charles Oxnard, have showed that they belonged to an ordinary ape species that became extinct and bore no resemblance to humans (Zuckerman 1970a , b ; Oxnard 1970 ).
The next stage of evolution is Homo , which is further divided into Homo habilis , Homo erectus and Homo sapiens . Each of these is considered to be one another’s ancestor. However recent fossil findings by the paleoanthropologist have revealed that Australopithecus , Homo habilis , and Homo erectus lived in different parts of the world at the same time (Walker 1980 ; Kesol 1970 ; Leakey 1971 ). And certain groups of Homo erectus lived until modern times. Homo sapiens and Neanderthals have also co-existed at the same time and also in the same region (Kluger 1996 ). The invalidity of this claim becomes more obvious when paleontologist fail to find any evolutionary trends in these so-called ancestors of man. Stephen Jay Gould a paleontologist from Harvard University explains this deadlock:
“What has become of our ladder if there are three coexisting lineages of hominids ( A. africanus , the robust australopithecines , and H. habilies ), none clearly derived from another? Moreover, none of the three display any evolutionary trends during their tenure on earth.”(Gould 1976 )
This claim was also backed by Lord Solly Zuckerman, who studied Australopithecus fossils for fifteen years, finally concluded that there is, in fact, no such family tree branching out from ape-like creatures to man.
He also formed a ‘spectrum of science’, in which sciences ranging from those he considered scientific to those he considered unscientific were put together. According to him, the most scientific, that is, depending on the concrete data-fields are chemistry and physics. After these are the biological sciences and then the social sciences. The most unscientific sciences which are at the far end of the spectrum include the extra-sensory perception, telepathy, sixth sense and finally human evolution. He explains this formation;
“We then move right off the register of objective truth into those fields of presumed biological science, like extrasensory perception or the interpretation of man’s fossil history, where to the faithful [evolutionist] anything is possible – and where the ardent believer [in evolution] is sometimes able to believe several contradictory things at the same time.”(Zuckerman 1970a , b )
Keeping all the arguments and counter arguments in mind with respect to the theory of man’s evolution, I shall conclude by quoting a few sentences from Harun Yahya’s book, ‘Fascism: The Bloody Ideology of Darwinism’ ,
“...the theory of evolution is a claim evidently at variance with scientific findings. The theory’s claim on the origin of life is inconsistent with science, the evolutionary mechanisms it proposes have no evolutionary power, and fossils demonstrate that the intermediate forms required by the theory never existed. So, it certainly follows that the theory of evolution should be pushed aside as an unscientific idea.”(Yahya 2002a , b , c )
Richard C. Lewontin who is a well-known geneticist and an evolutionist from Harvard University claims that he is first and foremost a materialist and then a scientist. He confesses;
“It is not that the methods and institutions of science somehow compel us to accept a material explanation of the phenomenal world, but, on the contrary, that we are forced by our a priori adherence to material causes to create an apparatus of investigation and a set of concepts that produce material explanations, no matter how counter-intuitive, no matter how mystifying to the uninitiated. Moreover, that materialism is absolute, so we cannot allow a Divine Foot in the door.”(Lewontin 1997 )
So, in short, the evolutionists who give materialist answers to the hundreds of questions that arise in the conscious thinking mind of the modern man today, are not only further creating confusions but have in a way failed to satisfy the logical and rational human mind. How can one believe that an unconscious matter can create life? How can one believe that matter created thousands and thousands of living things and living species with their own distinct attributes, qualities, and characteristics when scientist until today are not sure even how a small thing such as a simple cell can be formed? They know that it is formed when proteins come together, but how they come together, in what ratios and form a cell is a process that they have failed to understand (The Usborne Internet Linked Encyclopedia of World History, s.v., “human cell” 2000 ). For many years now engineers from around the world have been trying to make a three-dimensional television that can match the quality of the human eye. Yes, they have being successful in making a three-dimensional television screen, but you cannot watch it without putting on special glasses; moreover, it only creates artificial three dimension. Similarly ears, engineers have failed to produce a device that can ensure the same quality and clarity of sound that the human ear perceives. Another thing which is even more important than seeing and hearing abilities is the ‘consciousness’ that man has been blessed with (Yahya 2002a , b , c ). It is this consciousness that creates the major difference between man and all other living species. It is this that takes man one step ahead of all others. It is this ability that makes us flee from a fire, but we can go back in the same fire to save someone. It is this ability that helps us to understand and comprehend, that despite of the best of qualities given to us in this world, there are certain things that are still beyond our reach, control and comprehension. Even we humans have limitations, and this concept was well taken and understood even by early man since antiquity. He also knew that he had no control over the elements and there was some “Divine Force” somewhere, which had everything under its control. Hence it would not be wrong to presume here, that it was at this point in time around approximately 50,000 to 40,000 years ago, that the modern man entered the scene, and all the other species predating him were not actually ‘man’, or his ancestors. Hence, man was born a man with the best of qualities and a consciousness to understand the ‘Divine’ which has helped him not only to conquer but also to rule the world.
Change history
19 february 2019.
The Editor-in-Chief has retracted this article
Ager, A. Derek. 1976. The nature of the fossil record. Proceedings of the British Geological Association 87: 133.
Google Scholar
Bahn, G. Paul, ed. 2002a. The atlas of world archaeology . Oxfordshire: Andromeda. Publisher is Checkmark Books.
Bahn, Paul G., ed. 2002b. The Atlas of World Archaeology , 16. Oxfordshire: Andromeda.
Bahn, Paul G., ed. 2002c. The Atlas of World Archaeology , 22. Oxfordshire: Andromeda.
Bahn, Paul G., ed. 2002d. The Atlas of World Archaeology , 27. Oxfordshire: Andromeda.
Bahn, Paul G., ed. 2002e. The Atlas of World Archaeology , 28–29. Oxfordshire: Andromeda.
Braidwood, J. Robert. 1967. Prehistoric men . Toledo: Scott Foresman and Company.
Braidwood, Robert J. 1964a. Prehistoric men , 1–9. Chicago: Chicago History Museum.
Braidwood, Robert J. 1964b. Prehistoric men , 10–11. Chicago: Chicago History Museum.
Darwin, Charles. 1964. The origin of species: a facsimile of the first edition , 184. Boston: Harvard University Press.
Futuyma, J. Douglas. 1983. Science on trail , 197. New York: Pantheon Books.
Gould, J.S. 1976. Natural history . Vol. 85, 30. New York: American Museum of Natural History.
Grant, Micheal. 1952. Ancient history . London: First Harper TorchBook Edition.
Grant, Micheal. 1965. Ancient history , 5. New York: Harper and Row.
Haywood, John. 1997. Atlas of world history . Oxford: Sandcastle Books.
Haywood, John. 1989. n.d. The Atlas of Past Times , 12–13.
Ingold, Tim, ed. 1994a. Companion encyclopaedia of anthropology . New York: Routledge.
Ingold, Tim, ed. 1994b. Companion encyclopedia of anthropology , 87–88. New York: Routledge.
Ingold, Tim, ed. 1994c. Companion encyclopedia of anthropology , 88–92. New York: Routledge.
Kesol, J.A. 1970. Physical anthropology . 1st ed, 221. New York.
Kluger, Jeffrey. 1996. Not So Extinct After All: The Primitive Homo Erectus May Have Survived Long Enough To Coexist With Modern Humans. Time .
Leakey, M.D. 1971. Olduvai Gorge . Vol. 3, 272. Cambridge: Cambridge University Press.
Lewontin, Richard. 1997. The demon-haunted world , 28. The New York Review of Books.
Marwick, Arthur. 1970. The nature of history , 192–193. London: Palgrave Macmillan.
Oxnard, E. Charles. 1970. The place of australopithecines in human evolution: grounds of doubt. Nature 258 (New York: 389.
Article Google Scholar
Price, Douglas T., and Gary M. Feinman. 1993. Images of the Past . 2nd ed. California: Mayfield Publishing Company.
Price, T. Douglas, and Gary M. Feinman. 1997a. Images of the Past , 10. Houston: Mayfield Publishing.
Price, T. Douglas, and Gary M. Feinman. 1997b. Images of the Past , 16. Houston: Mayfield Publishing.
Price, T. Douglas, and Gary M. Feinman. 1997c. Images of the Past , 12. Houston: Mayfield.
Price, T. Douglas, and Gary M. Feinman. 1997d. Images of the Past , 39–40. Houston: Mayfield.
Price, T. Douglas, and Gary M. Feinman. 1997e. Images of the Past , 39. Houston: Mayfield.
Price, T. Douglas, and Gary M. Feinman. 1997f. Images of the Past , 41. Houston: Mayfield.
Schultz, A. Emily, and Robert H. Lavenda. 1998a. Anthropology- a perspective on the human condition . Belmont: Wadsworth.
Schultz, Emily A., and Robert H. Lavenda. 1998b. Anthropology- a perspective on the human condition , 128–152. California: Mayfield.
Schultz, Emily A., and Robert H. Lavenda. 1998c. Anthropology- a perspective on the human condition , 175. California: Mayfield.
Schultz, Emily A., and Robert H. Lavenda. 1998d. Anthropology- a perspective on the human condition , 176. California: Mayfield.
n.d. The New Encyclopedia Britannica . 15th ed, 995.
Jane Bingham, Fiona Chandler, Sam Taplin. 2000. The Usborne Internet-linked Encyclopedia of World History. In World History. Usborne. ISBN 0746041683, 9780746041680.
Walker, Alan. 1980. Science . Vol. 207, 1103.
Yahya, Harun. 2002a. Fascism: the bloody ideology of Darwinism . Turkey: Kultur.
Yahya, Harun. 2002b. Fascism: The Bloody Ideology of Darwinism , 234. Ankara: Global Publishing.
Yahya, Harun. 2002c. Fascism: The Bloody Ideology of Darwinism , 232–234. Turkey: Global Publishing.
Zuckerman, Solly. 1970a. Beyond the ivory tower , 75–94. New York: Toplinger Publications.
Zuckerman, Solly. 1970b. Beyond The Ivory Tower , 19. New York: Toplinger Publications.
Download references
Acknowledgements
To my PhD Co-Supervisor Dr. Musarrat Hasan.
Self funded.
Availability of data and materials
The datasets generated or analyzed during the current study is not publicly available due [REASON WHY DATA ARE NOT PUBLIC] but are available from the corresponding author on reasonable request.
Author information
Authors and affiliations.
Department of Visual Arts & Graphic Design, Institute of Visual Arts & Design, Lahore College for Women University, Lahore, Pakistan
You can also search for this author in PubMed Google Scholar
Contributions
I am the sole author of this manuscript. The author read and approved the final manuscript.
Corresponding author
Correspondence to Sarah Umer .
Ethics declarations
Author’s information.
Dr. Sarah Umer – Assistant Professor Institute of Visual Arts & Design at Lahore College for Women University, Lahore, Pakistan.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Competing interests.
The author declares that she has no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional information
The Editor-in-Chief has retracted this article [1], because it was published in error before the peer review process was completed. Further post publication peer review determined that the article is not suitable for publication in the International Journal of Anthropology and Ethnology. The author does not agree to this retraction.
1 Umer A brief history of human evolution: challenging Darwin’s claim International Journal of Anthropology and Ethnology (2018) 2:6 https://doi.org/10.1186/s41257-018-0014-2
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article.
Umer, S. RETRACTED ARTICLE: A brief history of human evolution: challenging Darwin’s claim. Int. j. anthropol. ethnol. 2 , 6 (2018). https://doi.org/10.1186/s41257-018-0014-2
Download citation
Received : 13 August 2018
Accepted : 26 September 2018
Published : 29 October 2018
DOI : https://doi.org/10.1186/s41257-018-0014-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Neanderthals

IMAGES
VIDEO
COMMENTS
Human Evolution: Theory and Progress. January 2014. DOI: 10.1007/978-1-4419-0465-2_642. In book: Encyclopedia of Global Archaeology (pp.3520-3532) Chapter: Human Evolution: Theory and Progress ...
The past, present and future of human evolution. María Martinón-Torres weighs up an original study on where we come from — and where we're going. A 1.8-million-year-old skull discovered in ...
1. Introduction: the big questions in modern human origins The first question which should be addressed in any discussion of the origin and evolution of Homo sapiens is which diagnosis of the species is going to be used. A paper using the classic multiregional concept of H. sapiens [1,2]
Man in light of the so-called history of human evolution. - but " also to understand that if the specie roaming the earth for a million years was truly man 's ancestor, as is claimed by Charles Darwin. Then what took man's ancestor so long to show signs of development that we only witness in the last 12000 years.
The new findings and their implications to the late stage of human evolution have been explored by different studies recently, all dedicated to some extent to understanding the trajectory of human evolution from the Chibanian to the present, focusing on specific topics like the origins of our species (Mounier and Mirazón Lahr, 2016, Mounier ...
The Journal of Human Evolution concentrates on publishing the highest quality papers covering all aspects of human evolution. The central focus is aimed jointly at paleoanthropological work, covering human and primate fossils, and at comparative studies of living species, including both …. View full aims & scope. $4170. Article publishing charge.
Introduction. In 1960, Jane Goodall established the first long-term field study of chimpanzees (Pan troglodytes) at what is now Gombe National Park, Tanzania, just over a century after On the origin of species (Darwin, 1859) laid the foundation for an evolutionary understanding of human origins.Goodall's mentor, Louis Leakey, hoped that studying living apes would shed light on the behaviour of ...
This Review focuses on recent advances in evolutionary genomics as they relate to our understanding of the origins and genetic basis for disease. Evolutionary medicine is a larger field that has ...
Understanding Human Evolution Human life, and how we came to be, is one of the greatest scienti c and philosophical questions of our time. This compact and accessible book pre-sents a modern view of human evolution. Written by a leading authority, it lucidly and engagingly explains not only the evolutionary process, but the
However, the recent discovery of modern human fossils in Greece and Israel dating to about 210 to 177 ka ago ( 9, 10) and ancient European genomes show that there were multiple out-of-Africa dispersals in the last 400,000 years, during which early humans and Neanderthals interbred ( 11, 12 ). Unlike what happened 60 ka ago ( 13 ), the offspring ...
Tennessen, J. A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64-69 (2012). This article represents one of the earliest ...
1. Introduction. Human evolution is characterized by speciation, extinction and dispersal events that have been linked to both global and/or regional palaeoclimate records [1-7].Many theories have been proposed to link environmental changes to these human evolution events [8-11].This synthesis paper presents each of these theories in the context of the pulsed climate variability conceptual ...
Evolutionary biology underwent several significant transformations in the period after 1930. While mid-century was dominated by. the evolutionary synthesis and the professionalization of ...
Uses an engaging and lucid style suitable for students entering the field. Follows the theme that the human anatomy is evolving and that it helps us understand our past. Includes current findings of genomics into understanding the more recent stages of human evolution. Part of the book series: Springer Texts in Social Sciences (STSS) 16k Accesses.
Research into the deep history of the human species is a relatively young science which can be divided into two broad periods. The first spans the century between the publication of Darwin's Origin and the end of World War II. This period is characterized by the recovery of the first non-modern human fossils and subsequent attempts at reconstructing family trees as visual representations of ...
Figure 1. Male and female human figures from the plaque of the Pioneer 10 and 11 spacecrafts. The pictorial message was intended to describe the origin of the probe for potential extraterrestrial life. It shows several typically human traits, such as bipedalism, nakedness, arched nose, large head, and opposable thumbs.
Darwin's theory of evolution through selection very well explains changes in time of heritable phenotypes. In the early 1900s, focusing on the evolution of genetic variants in the population, R. A. Fisher, S. Wright, and J. B. S. Haldane made fundamental theoretical contributions to population genetics (Provine 1971), Fisher in his 1922 paper (Fisher 1922), which was the first to introduce ...
evolution within the world itself, rather than in a c ontrolled laboratory. environment, which can shed light on how the many random (e.g., genetic. drift) and nonrandom (e.g., adaptati on ...
Abstract. Humans diverged from apes (chimpanzees, specifically) toward the end of the Miocene ~9.3 million to 6.5 million years ago. Understanding the origins of the human lineage (hominins) requires reconstructing the morphology, behavior, and environment of the chimpanzee-human last common ancestor.
Index. Download. XML. In this masterwork, Russell H. Tuttle synthesizes a vast research literature in primate evolution and behavior to explain how apes and humans evolved in relatio...
conceptions (ACs) in this paper. They are the views of young learners that differ significantly from the scientific conceptions described by scientists. Even teachers have been shown to possess ACs in ... human evolution Research Design This study aims to gather the qualitative views of grade 12 students on how human ancestors looked like ...
Smithsonian Human Origins Program & Field Research. The Smithsonian's Human Origins Program conducts field and lab research on the evolution of early human adaptations. Our key research partners are in East Africa and East Asia - especially in Kenya, China, and Indonesia. Our digs and studies in these regions, along with investigations by ...
Human evolution. Human evolution is the lengthy process of change by which people originated from apelike ancestors. Scientific evidence shows that the physical and behavioral traits shared by all people originated from apelike ancestors and evolved over a period of approximately six million years. One of the earliest defining human traits ...
Moreover, while keeping man's progress under consideration of the last 12000 years, it will further shed light on why there are serious reservations about Charles Darwin theory of human evolution. As many scientists, evolutionists, archeologist and different religious scriptures strongly claim that man came to the earth fully developed and ...